Computational Biology
Genomics, bioinformatics, and quantitative methods in biology
Genomics, bioinformatics, and quantitative methods in biology
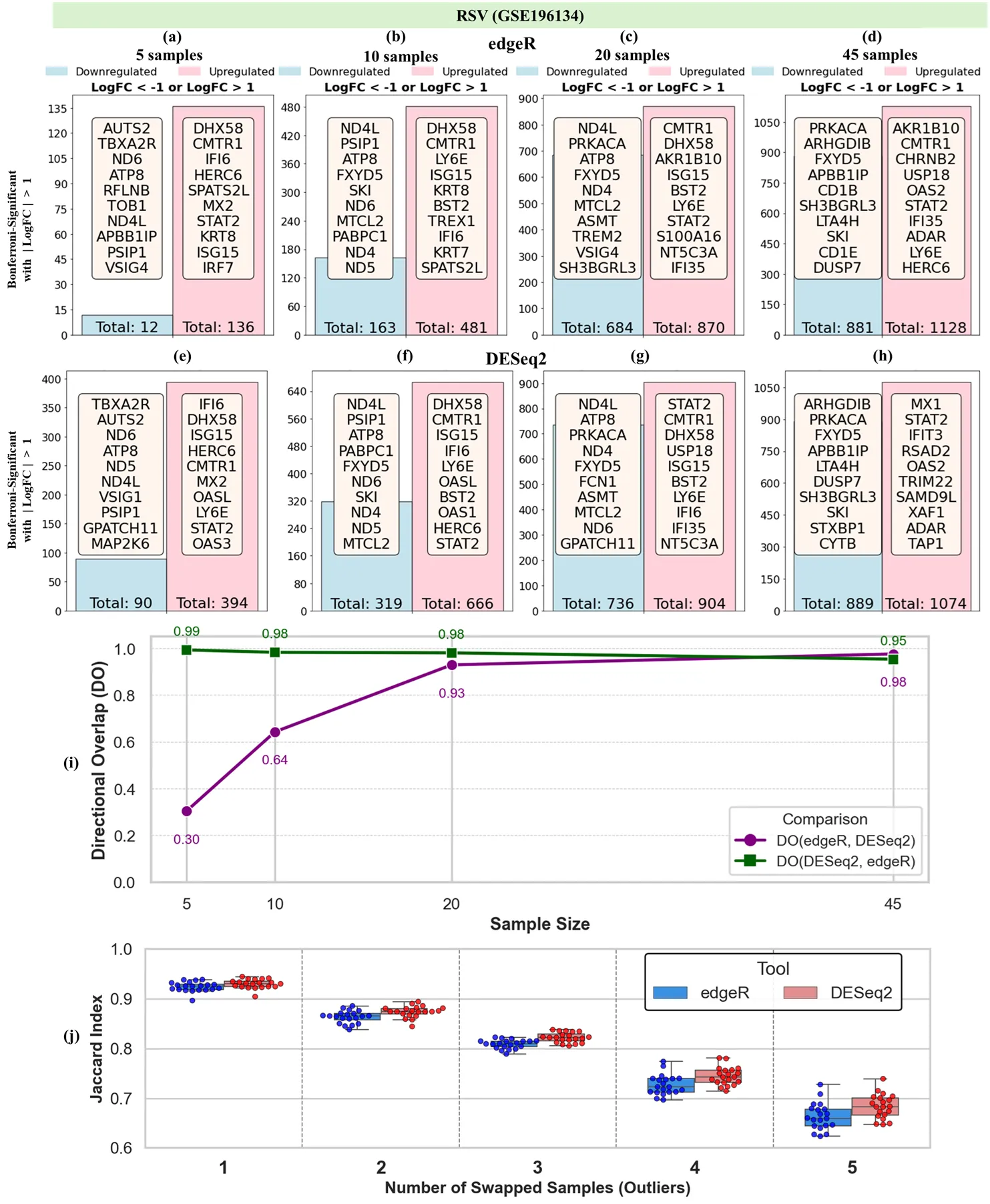
Differential gene expression (DGE) analysis is foundational to transcriptomic research, yet tool selection can substantially influence results. This study presents a comprehensive comparison of two widely used DGE tools, edgeR and DESeq2, using real and semi-simulated bulk RNA-Seq datasets spanning viral, bacterial, and fibrotic conditions. We evaluated tool performance across three key dimensions: (1) sensitivity to sample size and robustness to outliers; (2) classification performance of uniquely identified gene sets within the discovery dataset; and (3) generalizability of tool-specific gene sets across independent studies. First, both tools showed similar responses to simulated outliers, with Jaccard similarity between the DEG sets from perturbed and original (unperturbed) data decreasing as more outliers were added. Second, classification models trained on tool-specific genes showed that edgeR achieved higher F1 scores in 9 of 13 contrasts and more frequently reached perfect or near-perfect precision. Dolan-More performance profiles further indicated that edgeR maintained performance closer to optimal across a greater proportion of datasets. Third, in cross-study validation using four independent SARS-CoV-2 datasets, gene sets uniquely identified by edgeR yielded higher AUC, precision, and recall in classifying samples from held-out datasets. This pattern was consistent across folds, with some test cases achieving perfect separation using edgeR-specific genes. In contrast, DESeq2-specific genes showed lower and more variable performance across studies. Overall, our findings highlight that while DESeq2 may identify more DEGs even under stringent significance conditions, edgeR yields more robust and generalizable gene sets for downstream classification and cross-study replication, which underscores key trade-offs in tool selection for transcriptomic analyses.
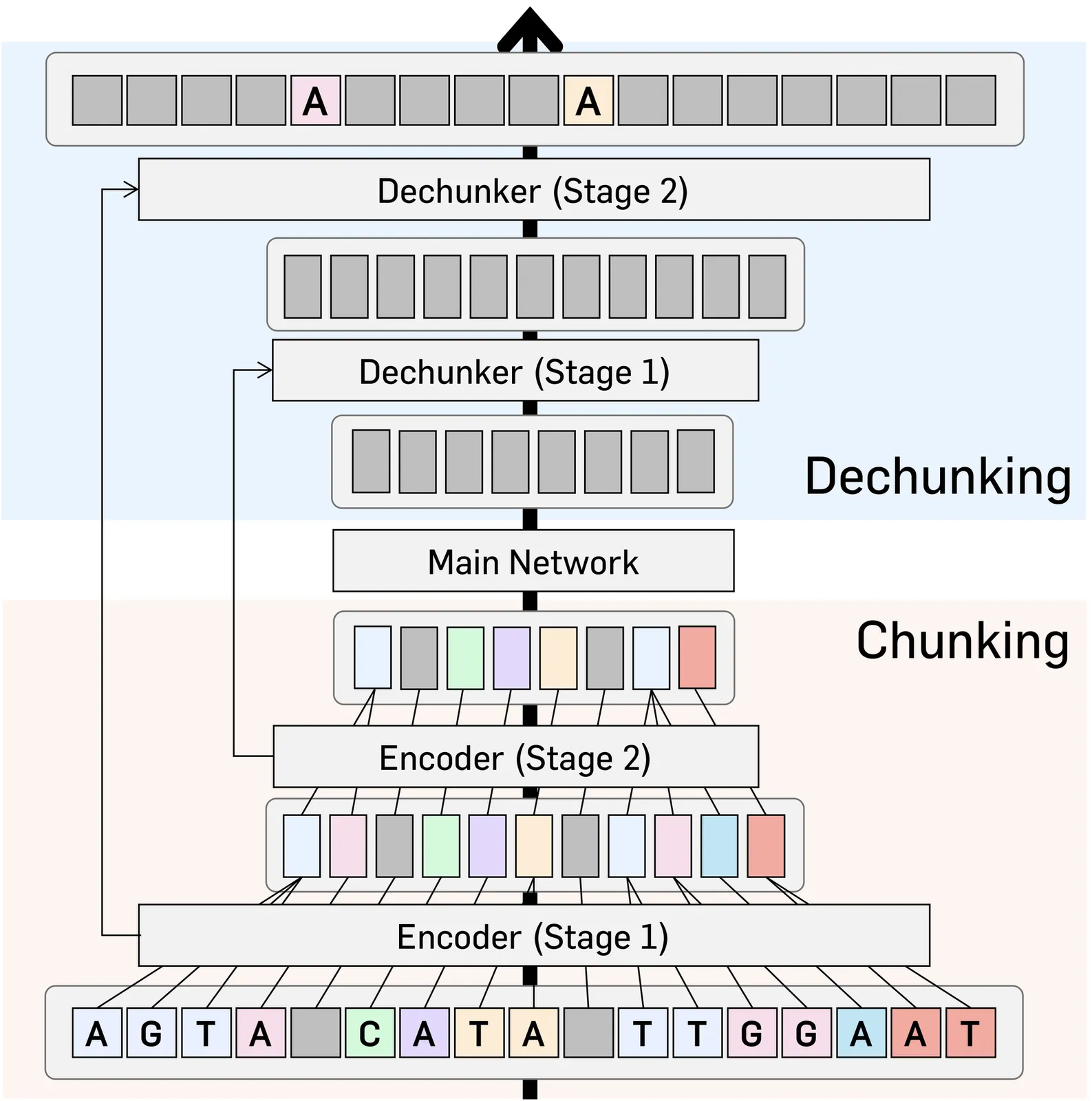
DNA language models have emerged as powerful tools for decoding the complex language of DNA sequences. However, the performance of these models is heavily affected by their tokenization strategy, i.e., a method used to parse DNA sequences into a shorter sequence of chunks. In this work, we propose DNACHUNKER, which integrates a learnable dynamic DNA tokenization mechanism and is trained as a masked language model. Adopting the dynamic chunking procedure proposed by H-Net, our model learns to segment sequences into variable-length chunks. This dynamic chunking offers two key advantages: it's resilient to shifts and mutations in the DNA, and it allocates more detail to important functional areas. We demonstrate the performance of DNACHUNKER by training it on the human reference genome (HG38) and testing it on the Nucleotide Transformer and Genomic benchmarks. Further ablative experiments reveal that DNACHUNKER learns tokenization that grasps biological grammar and uses smaller chunks to preserve detail in important functional elements such as promoters and exons, while using larger chunks for repetitive, redundant regions.
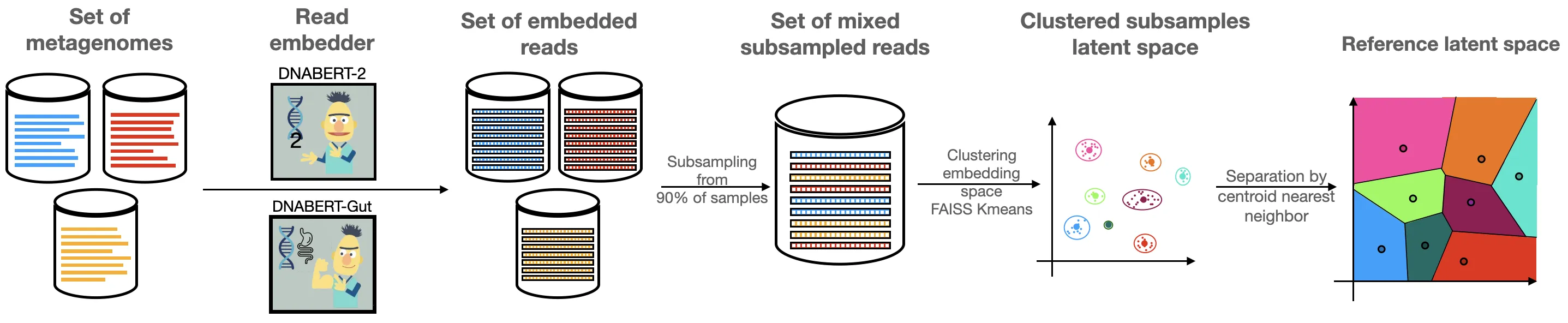
Metagenomic disease prediction commonly relies on species abundance tables derived from large, incomplete reference catalogs, constraining resolution and discarding valuable information contained in DNA reads. To overcome these limitations, we introduce MetagenBERT, a Transformer based framework that produces end to end metagenome embeddings directly from raw DNA sequences, without taxonomic or functional annotations. Reads are embedded using foundational genomic language models (DNABERT2 and the microbiome specialized DNABERTMS), then aggregated through a scalable clustering strategy based on FAISS accelerated KMeans. Each metagenome is represented as a cluster abundance vector summarizing the distribution of its embedded reads. We evaluate this approach on five benchmark gut microbiome datasets (Cirrhosis, T2D, Obesity, IBD, CRC). MetagenBERT achieves competitive or superior AUC performance relative to species abundance baselines across most tasks. Concatenating both representations further improves prediction, demonstrating complementarity between taxonomic and embedding derived signals. Clustering remains robust when applied to as little as 10% of reads, highlighting substantial redundancy in metagenomes and enabling major computational gains. We additionally introduce MetagenBERT Glob Mcardis, a cross cohort variant trained on the large, phenotypically diverse MetaCardis cohort and transferred to other datasets, retaining predictive signal including for unseen phenotypes, indicating the feasibility of a foundation model for metagenome representation. Robustness analyses (PERMANOVA, PERMDISP, entropy) show consistent separation of different states across subsamples. Overall, MetagenBERT provides a scalable, annotation free representation of metagenomes pointing toward future phenotype aware generalization across heterogeneous cohorts and sequencing technologies.
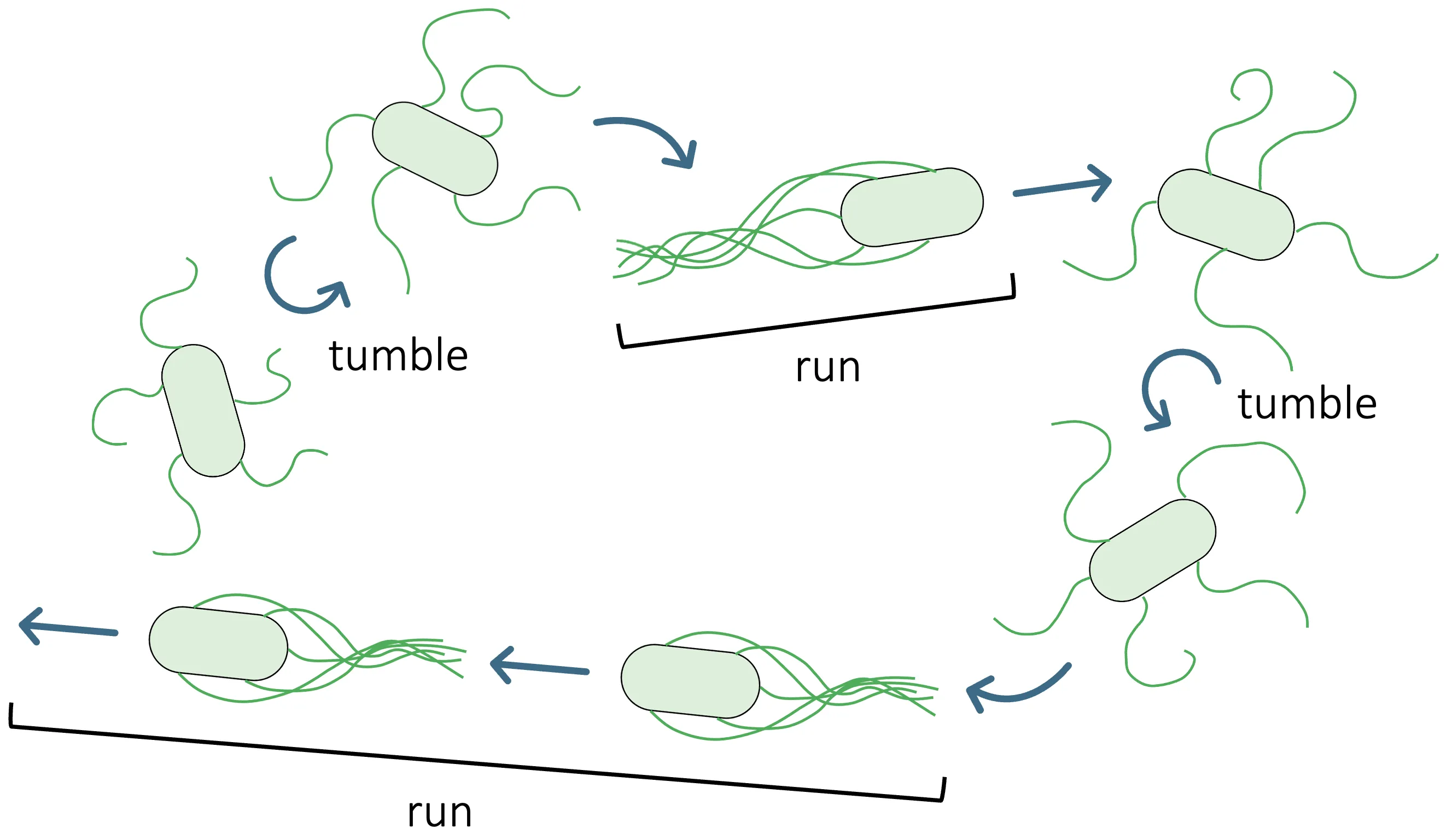
Bacterial chemotactic sensing converts noisy chemical signals into running and tumbling. We analyze the static sensing limits of mixed Tar/Tsr chemoreceptor clusters in individual Escherichia coli cells using a heterogeneous Monod-Wyman-Changeux (MWC) model. By sweeping a seven-dimensional parameter space, we compute three sensing performance metrics-channel capacity, effective Hill coefficient, and dynamic range. Across E. coli-like parameter regimes, we consistently observe pronounced local maxima of channel capacity, whereas neither the effective Hill coefficient nor the dynamic range exhibit comparable optimization. The capacity-achieving input distribution is bimodal, which implies that individual cells maximize information by sampling both low- and high concentration regimes. Together, these results suggest that, at the individual-cell level, channel capacity may be selected for in E. coli receptor clusters.
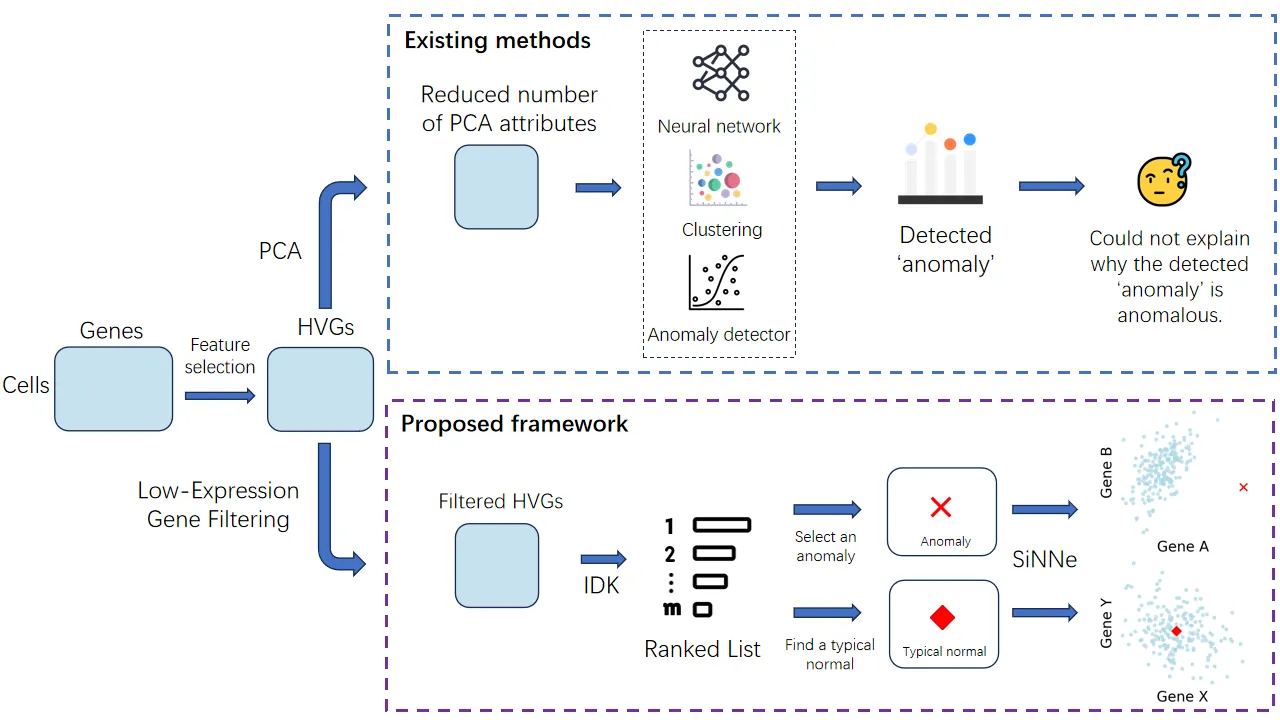
The detection of rare cell types in single-cell transcriptomics data is crucial for elucidating disease pathogenesis and tissue development dynamics. However, a critical gap that persists in current methods is their inability to provide an explanation based on genes for each cell they have detected as rare. We identify three primary sources of this deficiency. First, the anomaly detectors often function as "black boxes", designed to detect anomalies but unable to explain why a cell is anomalous. Second, the standard analytical framework hinders interpretability by relying on dimensionality reduction techniques, such as Principal Component Analysis (PCA), which transform meaningful gene expression data into abstract, uninterpretable features. Finally, existing explanation algorithms cannot be readily applied to this domain, as single-cell data is characterized by high dimensionality, noise, and substantial sparsity. To overcome these limitations, we introduce a framework for explainable anomaly detection in single-cell transcriptomics data which not only identifies individual anomalies, but also provides a visual explanation based on genes that makes an instance anomalous. This framework has two key ingredients that are not existed in current methods applied in this domain. First, it eliminates the PCA step which is deemed to be an essential component in previous studies. Second, it employs the state-of-art anomaly detector and explainer as the efficient and effective means to find each rare cell and the relevant gene subspace in order to provide explanations for each rare cell as well as the typical normal cell associated with the rare cell's closest normal cells.
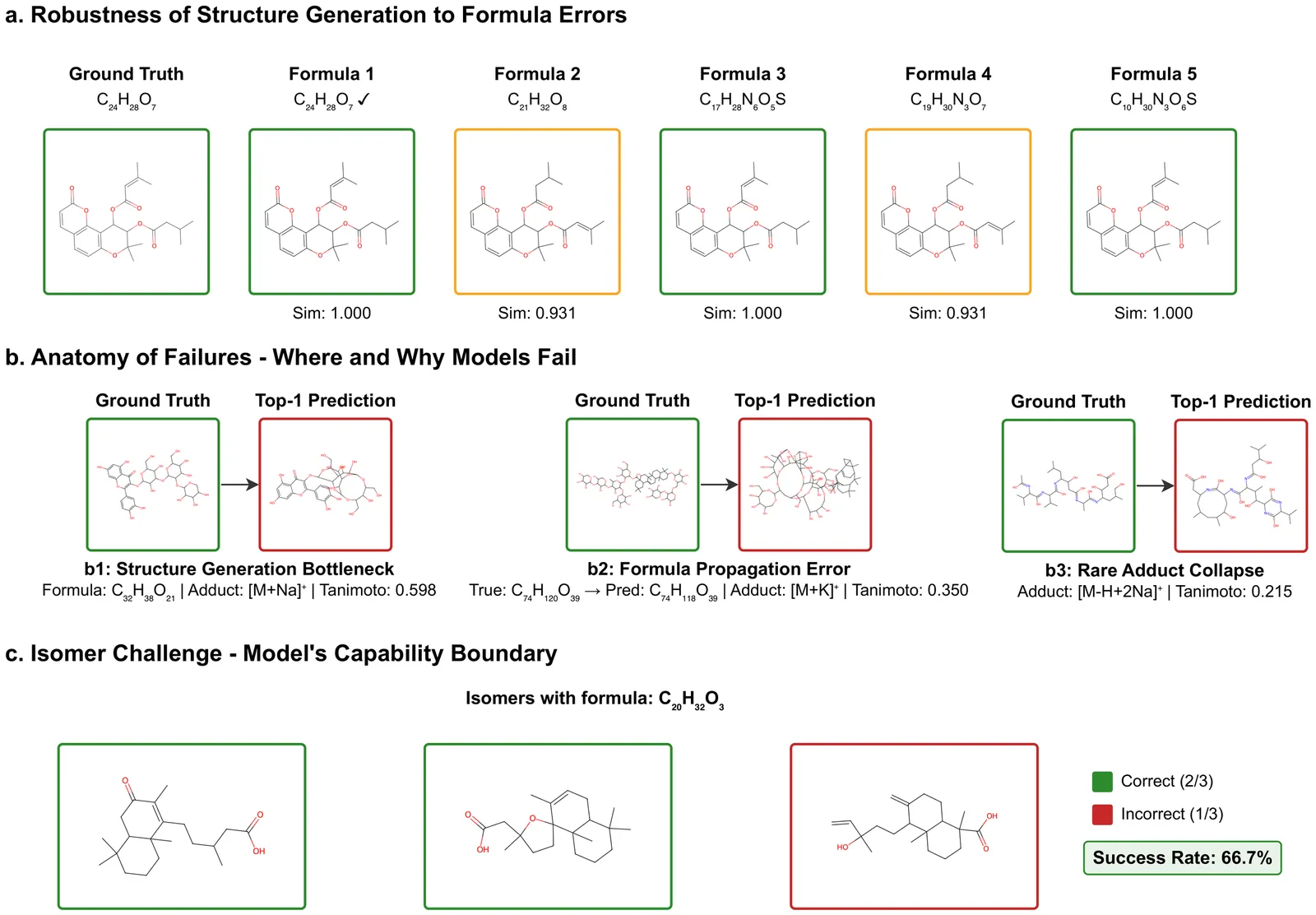
Liquid chromatography mass spectrometry (LC-MS)-based metabolomics and exposomics aim to measure detectable small molecules in biological samples. The results facilitate hypothesis-generating discovery of metabolic changes and disease mechanisms and provide information about environmental exposures and their effects on human health. Metabolomics and exposomics are made possible by the high resolving power of LC and high mass measurement accuracy of MS. However, a majority of the signals from such studies still cannot be identified or annotated using conventional library searching because existing spectral libraries are far from covering the vast chemical space captured by LC-MS/MS. To address this challenge and unleash the full potential of metabolomics and exposomics, a number of computational approaches have been developed to predict compounds based on tandem mass spectra. Published assessment of these approaches used different datasets and evaluation. To select prediction workflows for practical applications and identify areas for further improvements, we have carried out a systematic evaluation of the state-of-the-art prediction algorithms. Specifically, the accuracy of formula prediction and structure prediction was evaluated for different types of adducts. The resulting findings have established realistic performance baselines, identified critical bottlenecks, and provided guidance to further improve compound predictions based on MS.
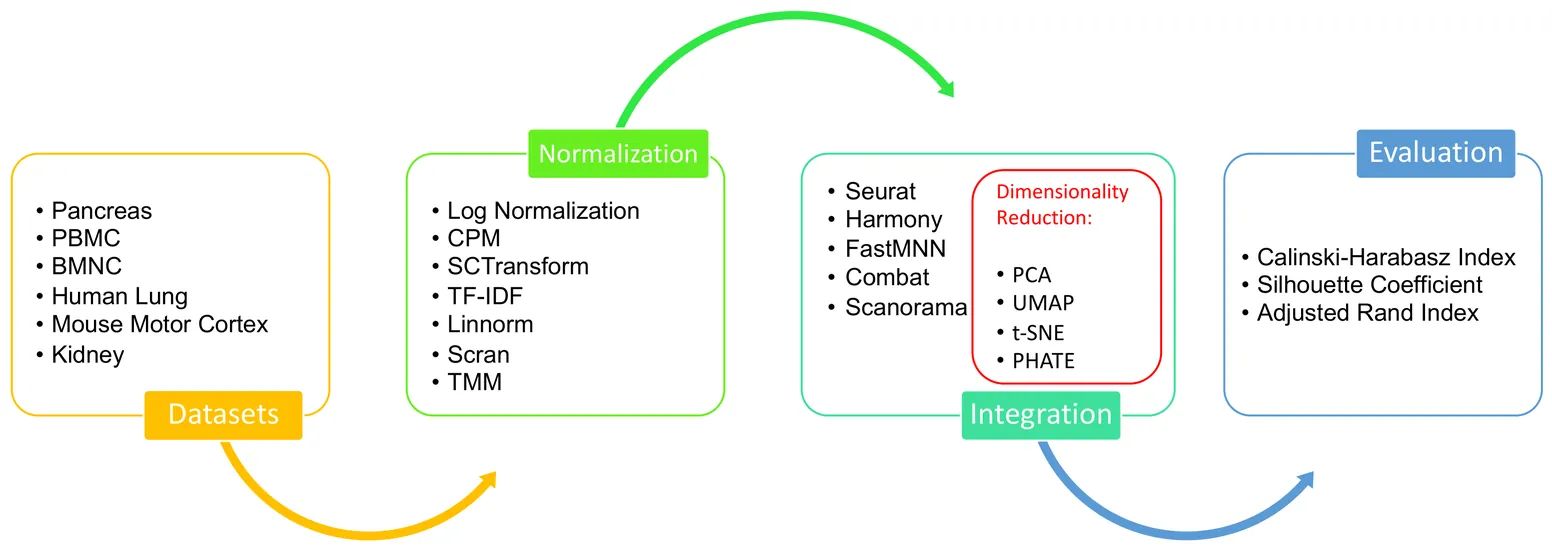
Single-cell data analysis has the potential to revolutionize personalized medicine by characterizing disease-associated molecular changes at the single-cell level. Advanced single-cell multimodal assays can now simultaneously measure various molecules (e.g., DNA, RNA, Protein) across hundreds of thousands of individual cells, providing a comprehensive molecular readout. A significant analytical challenge is integrating single-cell measurements across different modalities. Various methods have been developed to address this challenge, but there has been no systematic evaluation of these techniques with different preprocessing strategies. This study examines a general pipeline for single-cell data analysis, which includes normalization, data integration, and dimensionality reduction. The performance of different algorithm combinations often depends on the dataset sizes and characteristics. We evaluate six datasets across diverse modalities, tissues, and organisms using three metrics: Silhouette Coefficient Score, Adjusted Rand Index, and Calinski-Harabasz Index. Our experiments involve combinations of seven normalization methods, four dimensional reduction methods, and five integration methods. The results show that Seurat and Harmony excel in data integration, with Harmony being more time-efficient, especially for large datasets. UMAP is the most compatible dimensionality reduction method with the integration techniques, and the choice of normalization method varies depending on the integration method used.
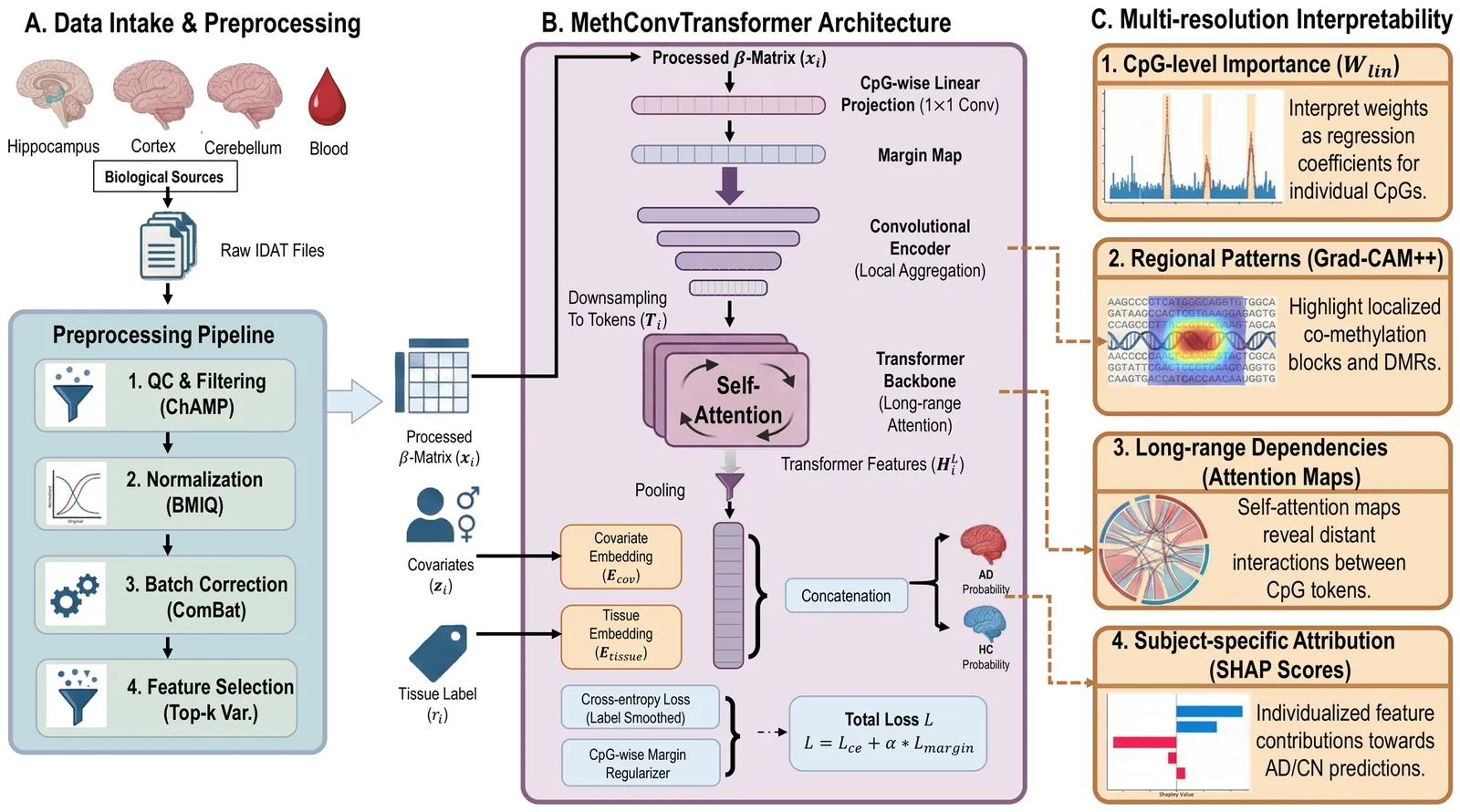
Alzheimer's disease (AD) is a multifactorial neurodegenerative disorder characterized by progressive cognitive decline and widespread epigenetic dysregulation in the brain. DNA methylation, as a stable yet dynamic epigenetic modification, holds promise as a noninvasive biomarker for early AD detection. However, methylation signatures vary substantially across tissues and studies, limiting reproducibility and translational utility. To address these challenges, we develop MethConvTransformer, a transformer-based deep learning framework that integrates DNA methylation profiles from both brain and peripheral tissues to enable biomarker discovery. The model couples a CpG-wise linear projection with convolutional and self-attention layers to capture local and long-range dependencies among CpG sites, while incorporating subject-level covariates and tissue embeddings to disentangle shared and region-specific methylation effects. In experiments across six GEO datasets and an independent ADNI validation cohort, our model consistently outperforms conventional machine-learning baselines, achieving superior discrimination and generalization. Moreover, interpretability analyses using linear projection, SHAP, and Grad-CAM++ reveal biologically meaningful methylation patterns aligned with AD-associated pathways, including immune receptor signaling, glycosylation, lipid metabolism, and endomembrane (ER/Golgi) organization. Together, these results indicate that MethConvTransformer delivers robust, cross-tissue epigenetic biomarkers for AD while providing multi-resolution interpretability, thereby advancing reproducible methylation-based diagnostics and offering testable hypotheses on disease mechanisms.
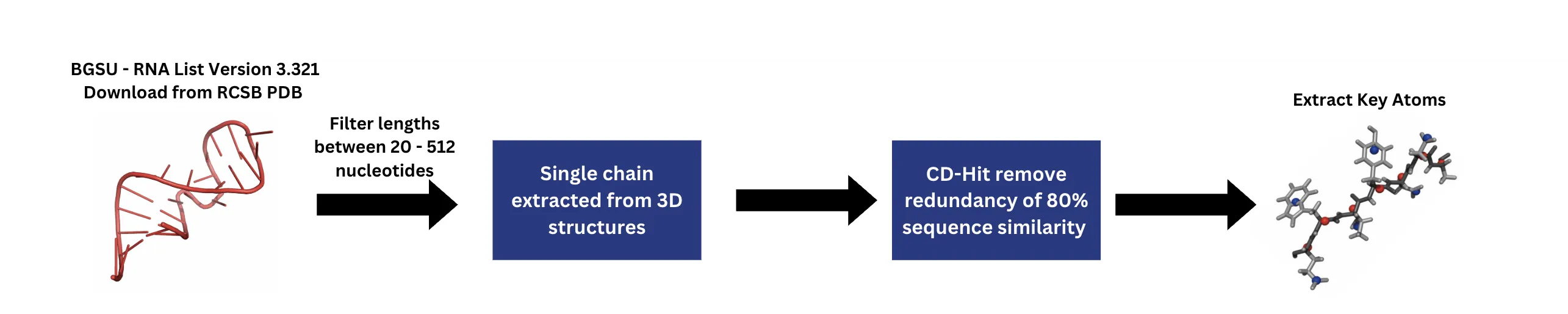
RNA's diverse biological functions stem from its structural versatility, yet accurately predicting and designing RNA sequences given a 3D conformation (inverse folding) remains a challenge. Here, I introduce a deep learning framework that integrates Geometric Vector Perceptron (GVP) layers with a Transformer architecture to enable end-to-end RNA design. I construct a dataset consisting of experimentally solved RNA 3D structures, filtered and deduplicated from the BGSU RNA list, and evaluate performance using both sequence recovery rate and TM-score to assess sequence and structural fidelity, respectively. On standard benchmarks and RNA-Puzzles, my model achieves state-of-the-art performance, with recovery and TM-scores of 0.481 and 0.332, surpassing existing methods across diverse RNA families and length scales. Masked family-level validation using Rfam annotations confirms strong generalization beyond seen families. Furthermore, inverse-folded sequences, when refolded using AlphaFold3, closely resemble native structures, highlighting the critical role of geometric features captured by GVP layers in enhancing Transformer-based RNA design.
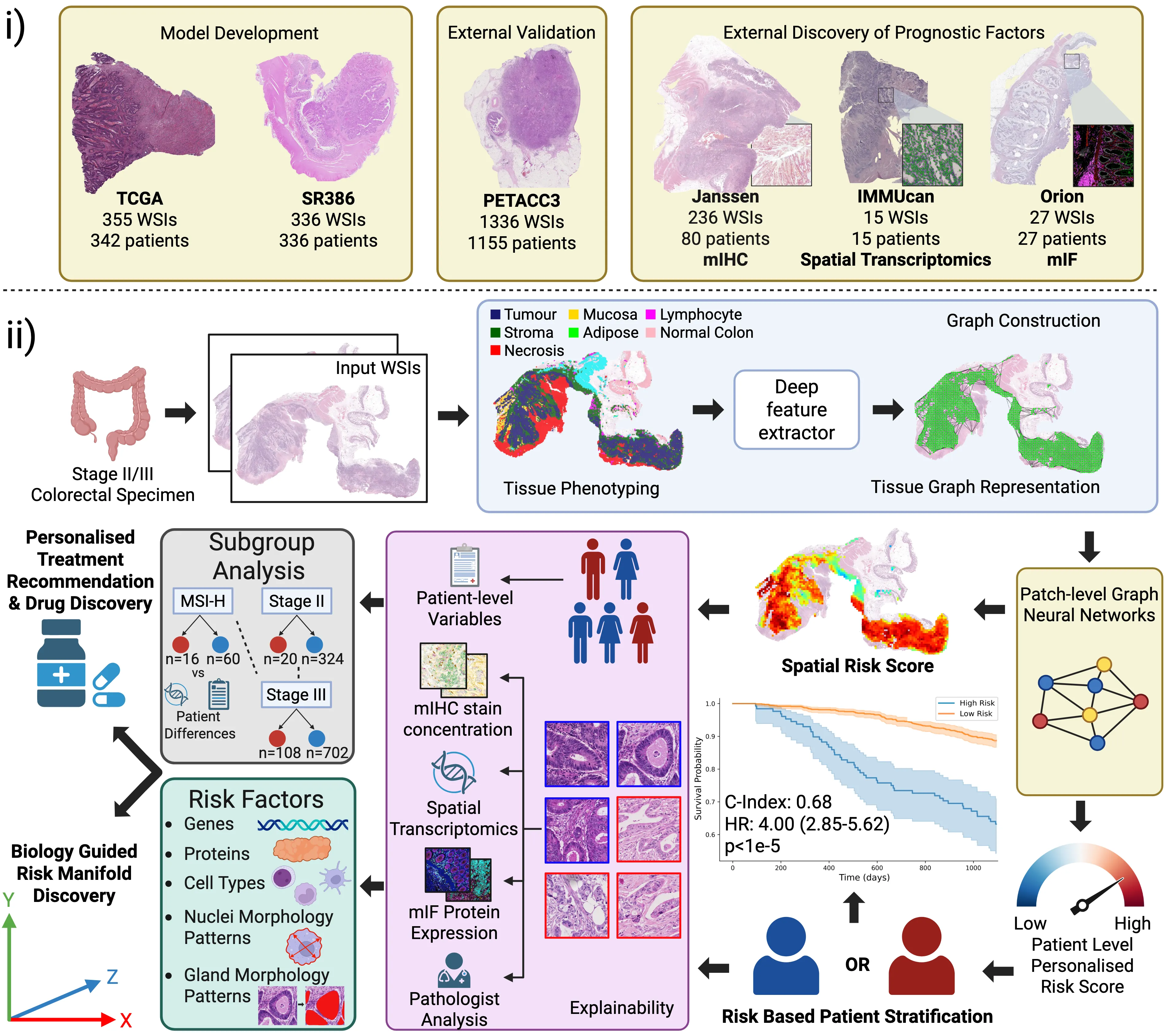
Routine histology contains rich prognostic information in stage II/III colorectal cancer, much of which is embedded in complex spatial tissue organisation. We present INSIGHT, a graph neural network that predicts survival directly from routine histology images. Trained and cross-validated on TCGA (n=342) and SURGEN (n=336), INSIGHT produces patient-level spatially resolved risk scores. Large independent validation showed superior prognostic performance compared with pTNM staging (C-index 0.68-0.69 vs 0.44-0.58). INSIGHT spatial risk maps recapitulated canonical prognostic histopathology and identified nuclear solidity and circularity as quantitative risk correlates. Integrating spatial risk with data-driven spatial transcriptomic signatures, spatial proteomics, bulk RNA-seq, and single-cell references revealed an epithelium-immune risk manifold capturing epithelial dedifferentiation and fetal programs, myeloid-driven stromal states including $\mathrm{SPP1}^{+}$ macrophages and $\mathrm{LAMP3}^{+}$ dendritic cells, and adaptive immune dysfunction. This analysis exposed patient-specific epithelial heterogeneity, stratification within MSI-High tumours, and high-risk routes of CDX2/HNF4A loss and CEACAM5/6-associated proliferative programs, highlighting coordinated therapeutic vulnerabilities.
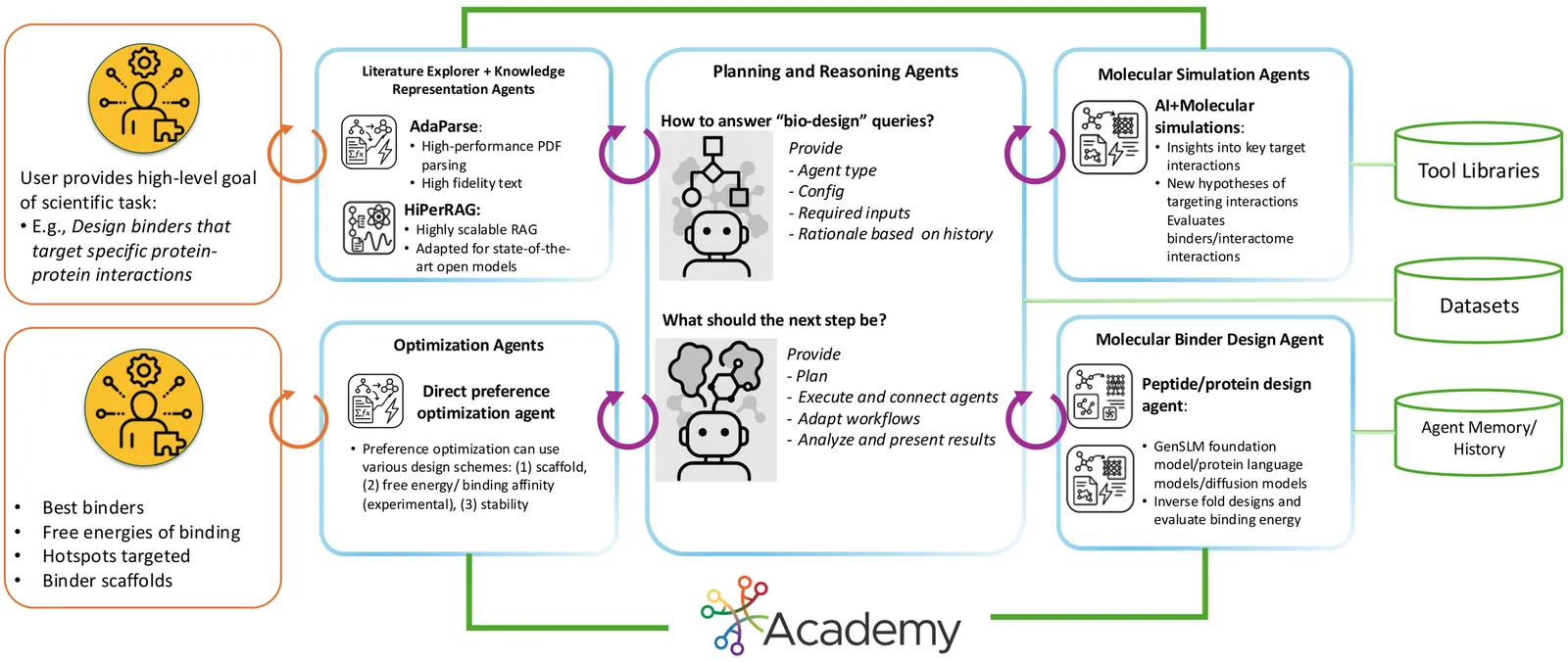
Intrinsically disordered proteins (IDPs) represent crucial therapeutic targets due to their significant role in disease -- approximately 80\% of cancer-related proteins contain long disordered regions -- but their lack of stable secondary/tertiary structures makes them "undruggable". While recent computational advances, such as diffusion models, can design high-affinity IDP binders, translating these to practical drug discovery requires autonomous systems capable of reasoning across complex conformational ensembles and orchestrating diverse computational tools at scale.To address this challenge, we designed and implemented StructBioReasoner, a scalable multi-agent system for designing biologics that can be used to target IDPs. StructBioReasoner employs a novel tournament-based reasoning framework where specialized agents compete to generate and refine therapeutic hypotheses, naturally distributing computational load for efficient exploration of the vast design space. Agents integrate domain knowledge with access to literature synthesis, AI-structure prediction, molecular simulations, and stability analysis, coordinating their execution on HPC infrastructure via an extensible federated agentic middleware, Academy. We benchmark StructBioReasoner across Der f 21 and NMNAT-2 and demonstrate that over 50\% of 787 designed and validated candidates for Der f 21 outperformed the human-designed reference binders from literature, in terms of improved binding free energy. For the more challenging NMNAT-2 protein, we identified three binding modes from 97,066 binders, including the well-studied NMNAT2:p53 interface. Thus, StructBioReasoner lays the groundwork for agentic reasoning systems for IDP therapeutic discovery on Exascale platforms.
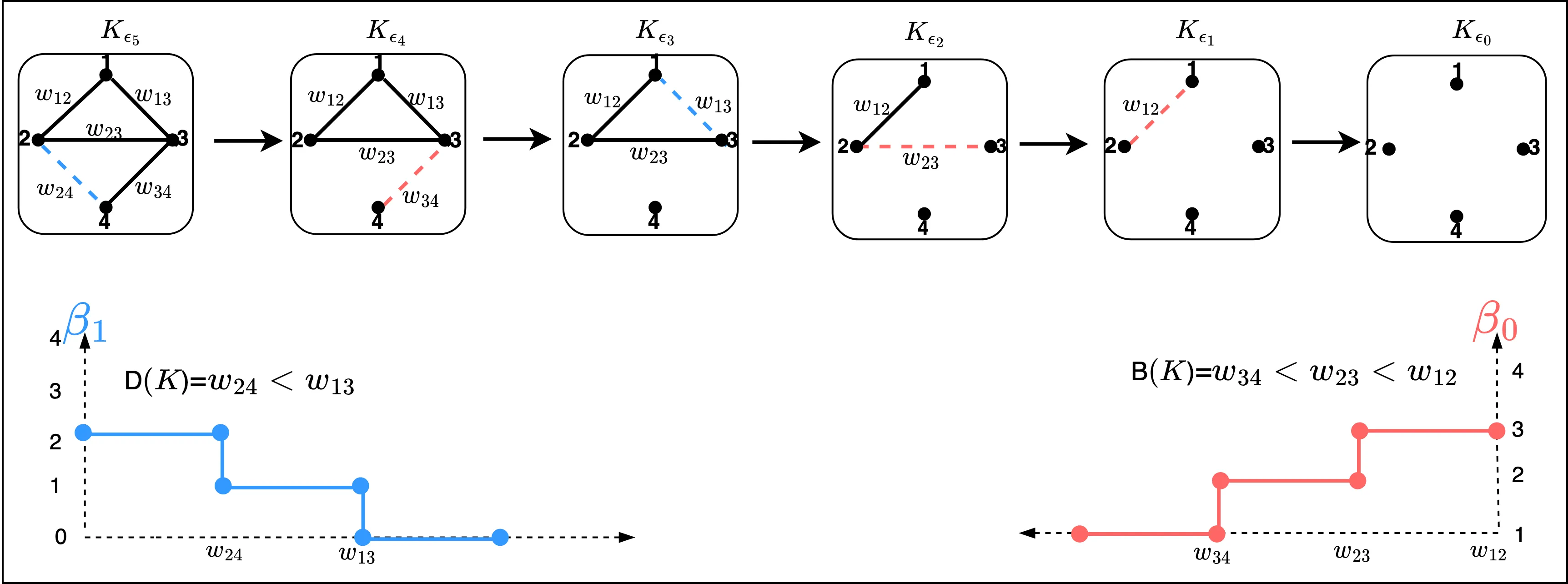
Identifying and comparing topological features, particularly cycles, across different topological objects remains a fundamental challenge in persistent homology and topological data analysis. This work introduces a novel framework for constructing cycle communities through two complementary approaches. First, a dendrogram-based methodology leverages merge-tree algorithms to construct hierarchical representations of homology classes from persistence intervals. The Wasserstein distance on merge trees is introduced as a metric for comparing dendrograms, establishing connections to hierarchical clustering frameworks. Through simulation studies, the discriminative power of dendrogram representations for identifying cycle communities is demonstrated. Second, an extension of Stratified Gradient Sampling simultaneously learns multiple filter functions that yield cycle barycenter functions capable of faithfully reconstructing distinct sets of cycles. The set of cycles each filter function can reconstruct constitutes cycle communities that are non-overlapping and partition the space of all cycles. Together, these approaches transform the problem of cycle matching into both a hierarchical clustering and topological optimization framework, providing principled methods to identify similar topological structures both within and across groups of topological objects.
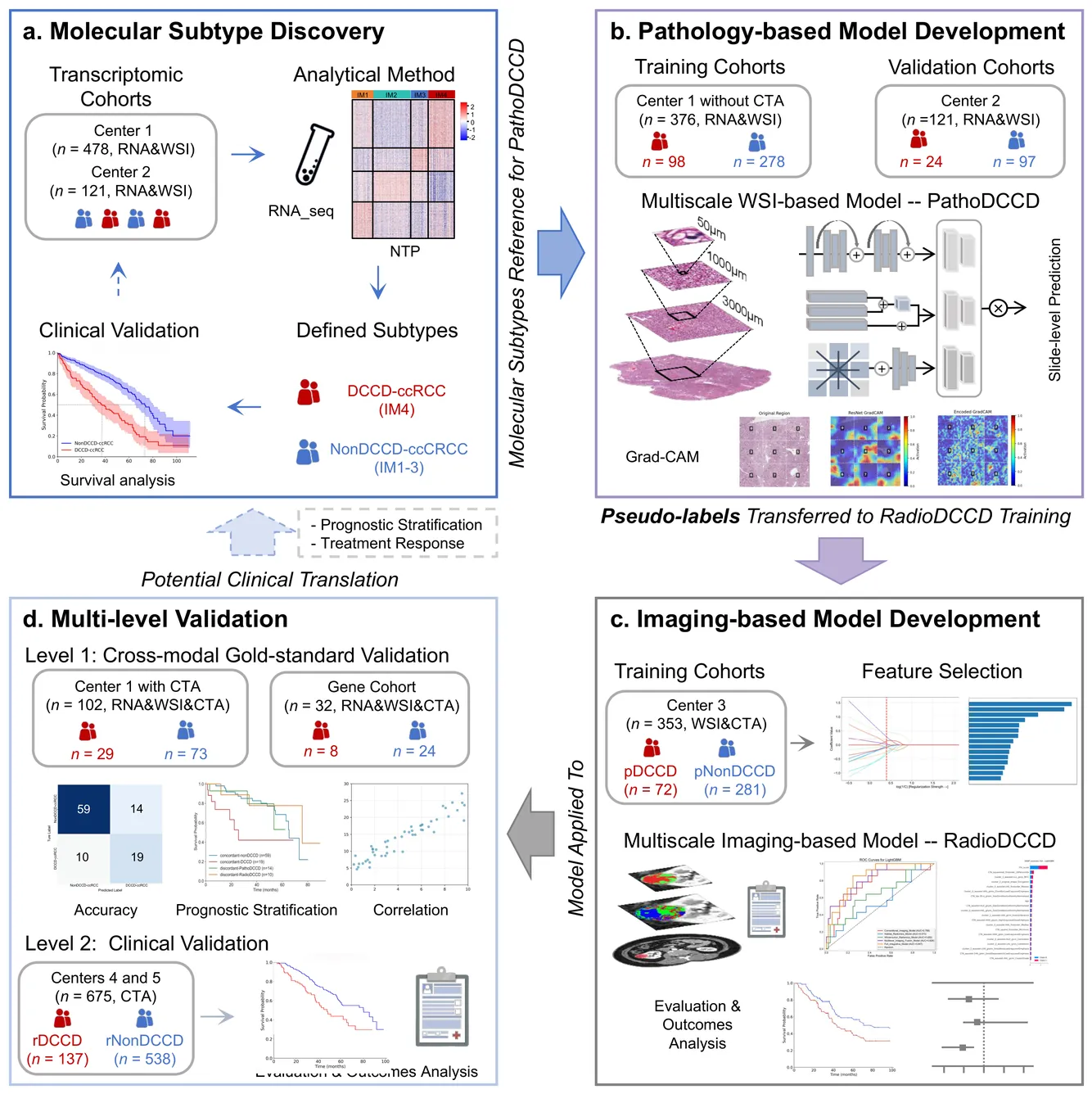
Clear cell renal cell carcinoma (ccRCC) exhibits extensive intratumoral heterogeneity on multiple biological scales, contributing to variable clinical outcomes and limiting the effectiveness of conventional TNM staging, which highlights the urgent need for multiscale integrative analytic frameworks. The lipid-deficient de-clear cell differentiated (DCCD) ccRCC subtype, defined by multi-omics analyses, is associated with adverse outcomes even in early-stage disease. Here, we establish a hierarchical cross-scale framework for the preoperative identification of DCCD-ccRCC. At the highest layer, cross-modal mapping transferred molecular signatures to histological and CT phenotypes, establishing a molecular-to-pathology-to-radiology supervisory bridge. Within this framework, each modality-specific model is designed to mirror the inherent hierarchical structure of tumor biology. PathoDCCD captured multi-scale microscopic features, from cellular morphology and tissue architecture to meso-regional organization. RadioDCCD integrated complementary macroscopic information by combining whole-tumor and its habitat-subregions radiomics with a 2D maximal-section heterogeneity metric. These nested models enabled integrated molecular subtype prediction and clinical risk stratification. Across five cohorts totaling 1,659 patients, PathoDCCD reliably recapitulated molecular subtypes, while RadioDCCD provided reliable preoperative prediction. The consistent predictions identified patients with the poorest clinical outcomes. This cross-scale paradigm unifies molecular biology, computational pathology, and quantitative radiology into a biologically grounded strategy for preoperative noninvasive molecular phenotyping of ccRCC.
 2512.12272
2512.12272Post-translational modifications (PTMs) serve as a dynamic chemical language regulating protein function, yet current proteomic methods remain blind to a vast portion of the modified proteome. Standard database search algorithms suffer from a combinatorial explosion of search spaces, limiting the identification of uncharacterized or complex modifications. Here we introduce OmniNovo, a unified deep learning framework for reference-free sequencing of unmodified and modified peptides directly from tandem mass spectra. Unlike existing tools restricted to specific modification types, OmniNovo learns universal fragmentation rules to decipher diverse PTMs within a single coherent model. By integrating a mass-constrained decoding algorithm with rigorous false discovery rate estimation, OmniNovo achieves state-of-the-art accuracy, identifying 51\% more peptides than standard approaches at a 1\% false discovery rate. Crucially, the model generalizes to biological sites unseen during training, illuminating the dark matter of the proteome and enabling unbiased comprehensive analysis of cellular regulation.
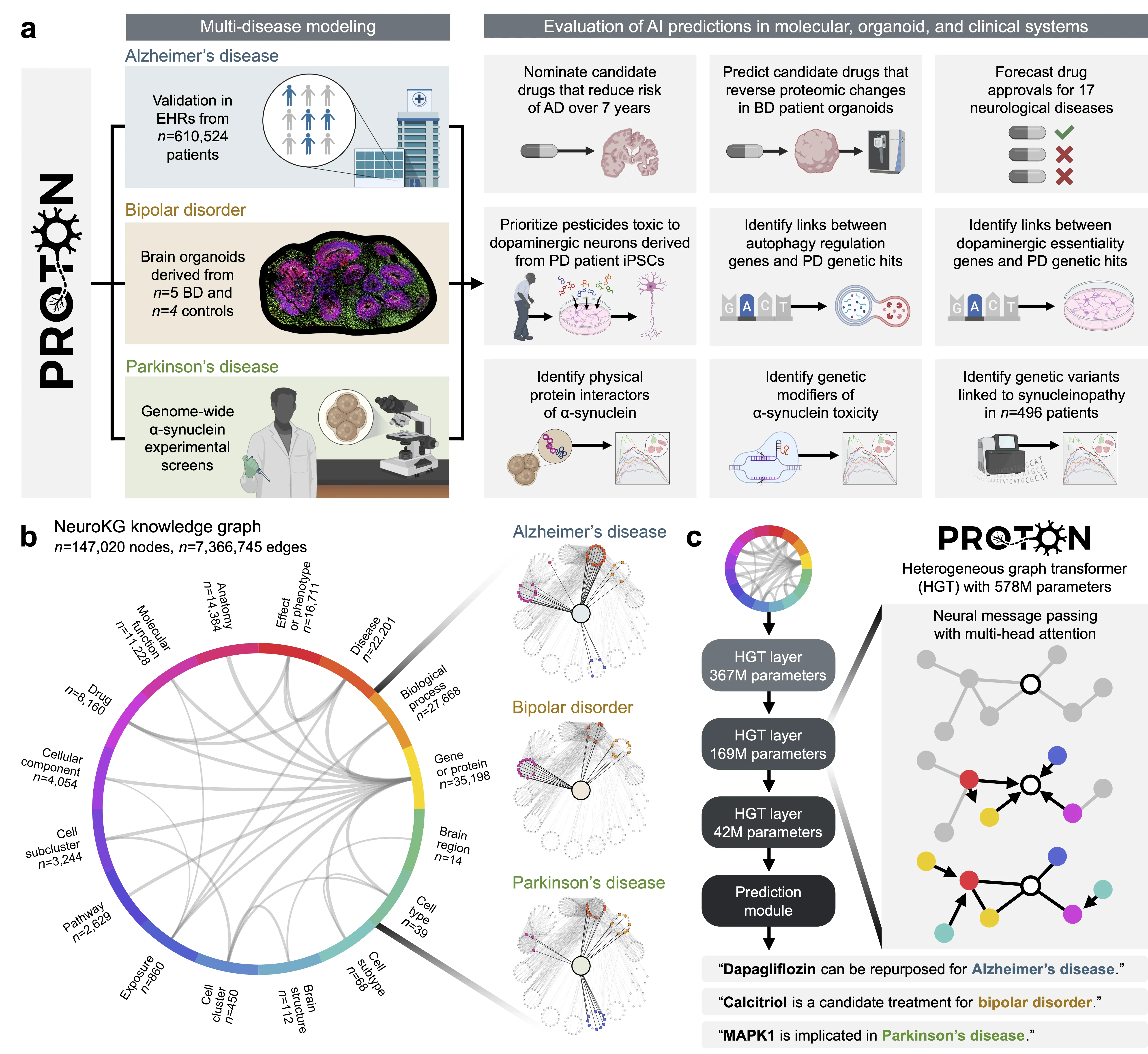
Neurological diseases are the leading global cause of disability, yet most lack disease-modifying treatments. We present PROTON, a heterogeneous graph transformer that generates testable hypotheses across molecular, organoid, and clinical systems. To evaluate PROTON, we apply it to Parkinson's disease (PD), bipolar disorder (BD), and Alzheimer's disease (AD). In PD, PROTON linked genetic risk loci to genes essential for dopaminergic neuron survival and predicted pesticides toxic to patient-derived neurons, including the insecticide endosulfan, which ranked within the top 1.29% of predictions. In silico screens performed by PROTON reproduced six genome-wide $α$-synuclein experiments, including a split-ubiquitin yeast two-hybrid system (normalized enrichment score [NES] = 2.30, FDR-adjusted $p < 1 \times 10^{-4}$), an ascorbate peroxidase proximity labeling assay (NES = 2.16, FDR $< 1 \times 10^{-4}$), and a high-depth targeted exome sequencing study in 496 synucleinopathy patients (NES = 2.13, FDR $< 1 \times 10^{-4}$). In BD, PROTON predicted calcitriol as a candidate drug that reversed proteomic alterations observed in cortical organoids derived from BD patients. In AD, we evaluated PROTON predictions in health records from $n = 610,524$ patients at Mass General Brigham, confirming that five PROTON-predicted drugs were associated with reduced seven-year dementia risk (minimum hazard ratio = 0.63, 95% CI: 0.53-0.75, $p < 1 \times 10^{-7}$). PROTON generated neurological hypotheses that were evaluated across molecular, organoid, and clinical systems, defining a path for AI-driven discovery in neurological disease.
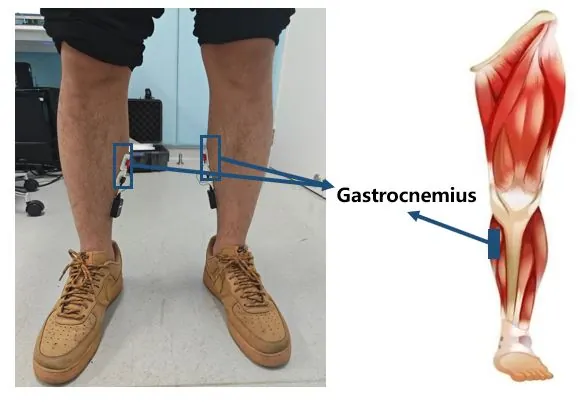
Rehabilitation exoskeletons have shown promising results in promoting recovery for stroke patients. Accurately and timely identifying the motion intentions of patients is a critical challenge in enhancing active participation during lower limb exoskeleton-assisted rehabilitation training. This paper proposes a Dual-Channel Attentive Fusion Network (DCAF-Net) that synergistically integrates pre-movement surface electromyography (sEMG) and inertial measurement unit (IMU) data for lower limb intention prediction in stroke patients. First, a dual-channel adaptive channel attention module is designed to extract discriminative features from 48 time-domain and frequency-domain features derived from bilateral gastrocnemius sEMG signals. Second, an IMU encoder combining convolutional neural network (CNN) and attention-based long short-term memory (attention-LSTM) layers is designed to decode temporal-spatial movement patterns. Third, the sEMG and IMU features are fused through concatenation to enable accurate recognition of motion intention. Extensive experiment on 11 participants (8 stroke subjects and 3 healthy subjects) demonstrate the effectiveness of DCAF-Net. It achieved a prediction accuracies of 97.19% for patients and 93.56% for healthy subjects. This study provides a viable solution for implementing intention-driven human-in-the-loop assistance control in clinical rehabilitation robotics.
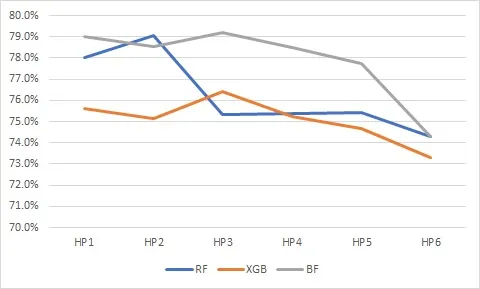
Cancer patients may undergo lengthy and painful chemotherapy treatments, comprising several successive regimens or plans. Treatment inefficacy and other adverse events can lead to discontinuation (or failure) of these plans, or prematurely changing them, which results in a significant amount of physical, financial, and emotional toxicity to the patients and their families. In this work, we build treatment failure models based on the Real World Evidence (RWE) gathered from patients' profiles available in our oncology EMR/EHR system. We also describe our feature engineering pipeline, experimental methods, and valuable insights obtained about treatment failures from trained models. We report our findings on five primary cancer types with the most frequent treatment failures (or discontinuations) to build unique and novel feature vectors from the clinical notes, diagnoses, and medications that are available in our oncology EMR. After following a novel three axes - performance, complexity and explainability - design exploration framework, boosted random forests are selected because they provide a baseline accuracy of 80% and an F1 score of 75%, with reduced model complexity, thus making them more interpretable to and usable by oncologists.
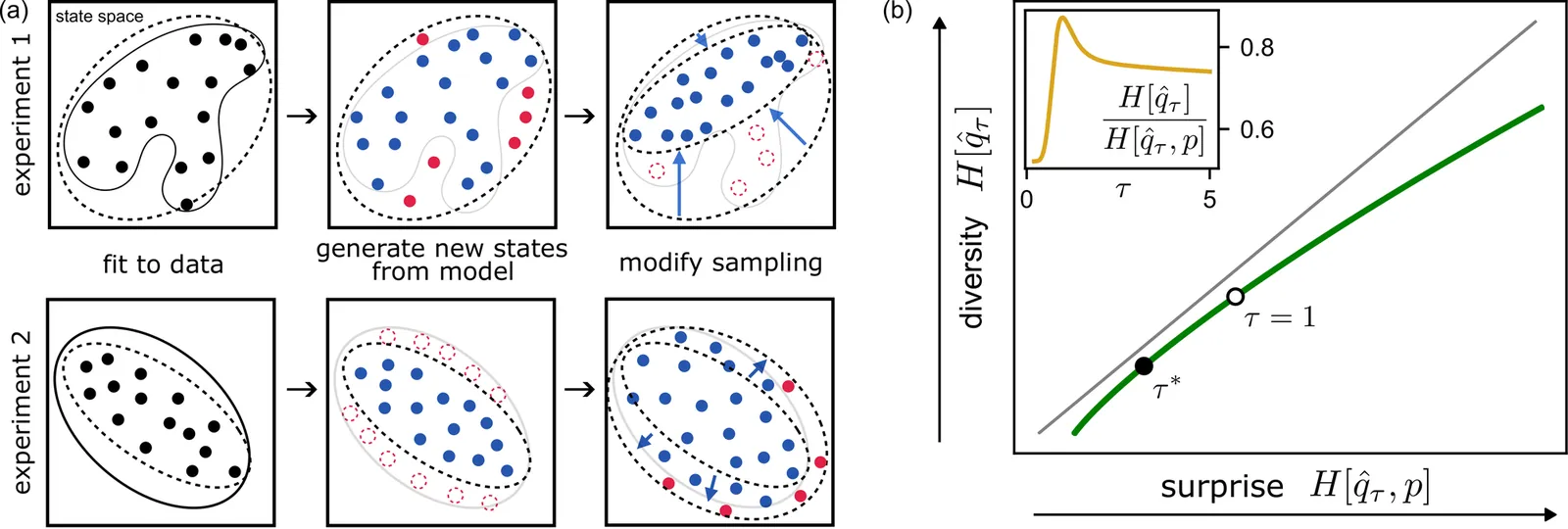
Generative models of complex systems often require post-hoc parameter adjustments to produce useful outputs. For example, energy-based models for protein design are sampled at an artificially low ''temperature'' to generate novel, functional sequences. This temperature tuning is a common yet poorly understood heuristic used across machine learning contexts to control the trade-off between generative fidelity and diversity. Here, we develop an interpretable, physically motivated framework to explain this phenomenon. We demonstrate that in systems with a large ''energy gap'' - separating a small fraction of meaningful states from a vast space of unrealistic states - learning from sparse data causes models to systematically overestimate high-energy state probabilities, a bias that lowering the sampling temperature corrects. More generally, we characterize how the optimal sampling temperature depends on the interplay between data size and the system's underlying energy landscape. Crucially, our results show that lowering the sampling temperature is not always desirable; we identify the conditions where \emph{raising} it results in better generative performance. Our framework thus casts post-hoc temperature tuning as a diagnostic tool that reveals properties of the true data distribution and the limits of the learned model.
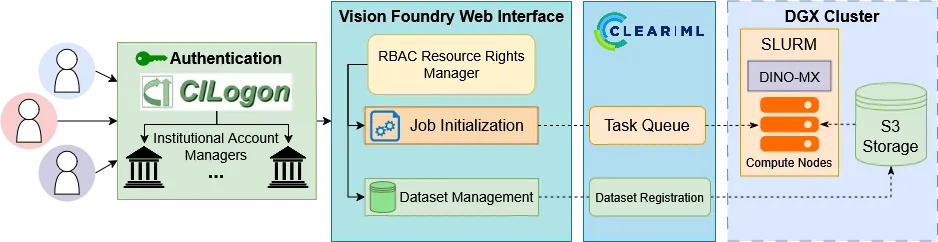
Self-supervised learning (SSL) leverages vast unannotated medical datasets, yet steep technical barriers limit adoption by clinical researchers. We introduce Vision Foundry, a code-free, HIPAA-compliant platform that democratizes pre-training, adaptation, and deployment of foundational vision models. The system integrates the DINO-MX framework, abstracting distributed infrastructure complexities while implementing specialized strategies like Magnification-Aware Distillation (MAD) and Parameter-Efficient Fine-Tuning (PEFT). We validate the platform across domains, including neuropathology segmentation, lung cellularity estimation, and coronary calcium scoring. Our experiments demonstrate that models trained via Vision Foundry significantly outperform generic baselines in segmentation fidelity and regression accuracy, while exhibiting robust zero-shot generalization across imaging protocols. By bridging the gap between advanced representation learning and practical application, Vision Foundry enables domain experts to develop state-of-the-art clinical AI tools with minimal annotation overhead, shifting focus from engineering optimization to clinical discovery.
In multicellular organisms, cells coordinate their activities through cell-cell communication (CCC), which are crucial for development, tissue homeostasis, and disease progression. Recent advances in single-cell and spatial omics technologies provide unprecedented opportunities to systematically infer and analyze CCC from these omics data, either by integrating prior knowledge of ligand-receptor interactions (LRIs) or through de novo approaches. A variety of computational methods have been developed, focusing on methodological innovations, accurate modeling of complex signaling mechanisms, and investigation of broader biological questions. These advances have greatly enhanced our ability to analyze CCC and generate biological hypotheses. Here, we introduce the biological mechanisms and modeling strategies of CCC, and provide a focused overview of more than 140 computational methods for inferring CCC from single-cell and spatial transcriptomic data, emphasizing the diversity in methodological frameworks and biological questions. Finally, we discuss the current challenges and future opportunities in this rapidly evolving field.