Image & Video Processing
Image processing, video analysis, and computational imaging
Image processing, video analysis, and computational imaging
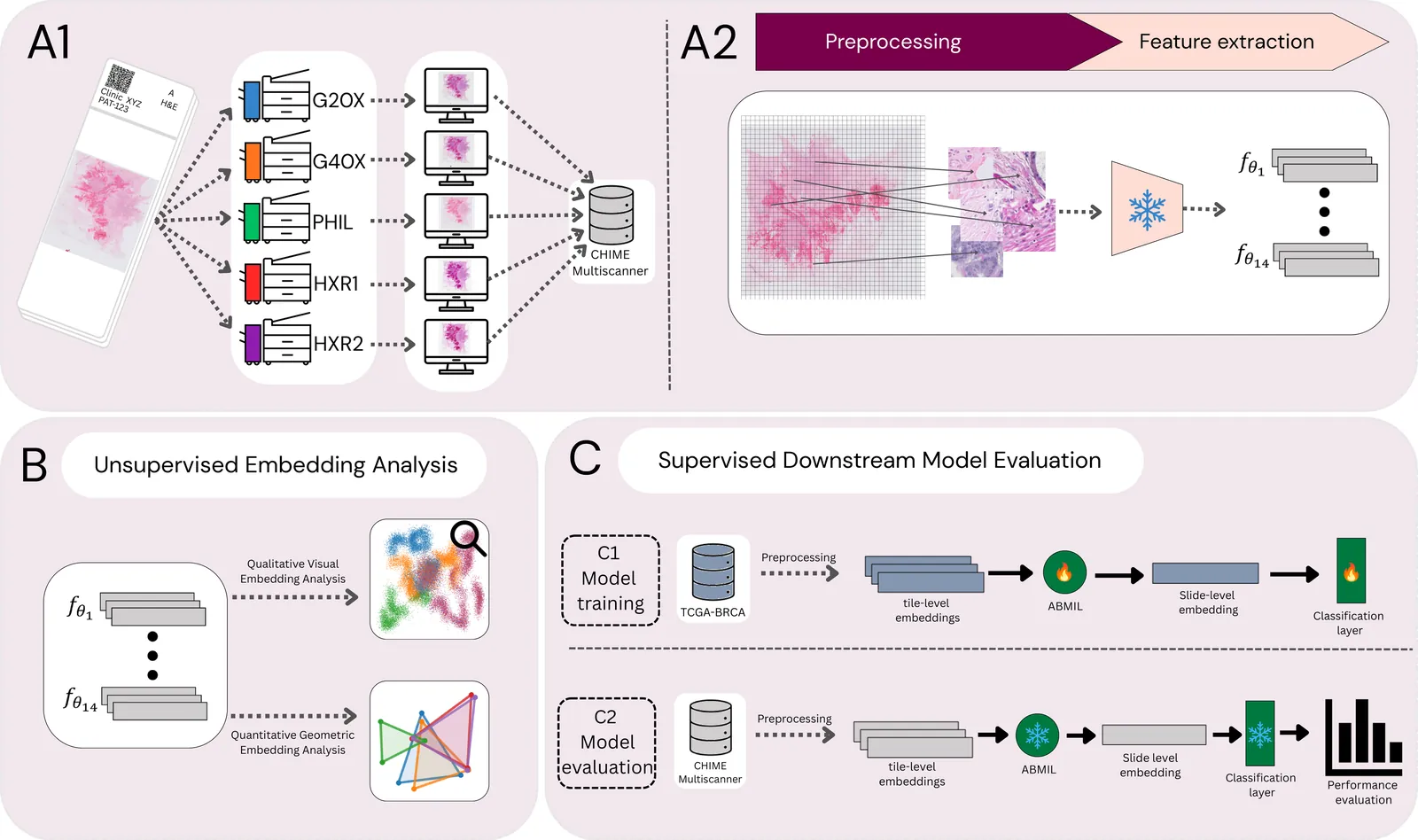
Pathology foundation models (PFMs) have become central to computational pathology, aiming to offer general encoders for feature extraction from whole-slide images (WSIs). Despite strong benchmark performance, PFM robustness to real-world technical domain shifts, such as variability from whole-slide scanner devices, remains poorly understood. We systematically evaluated the robustness of 14 PFMs to scanner-induced variability, including state-of-the-art models, earlier self-supervised models, and a baseline trained on natural images. Using a multiscanner dataset of 384 breast cancer WSIs scanned on five devices, we isolated scanner effects independently from biological and laboratory confounders. Robustness is assessed via complementary unsupervised embedding analyses and a set of clinicopathological supervised prediction tasks. Our results demonstrate that current PFMs are not invariant to scanner-induced domain shifts. Most models encode pronounced scanner-specific variability in their embedding spaces. While AUC often remains stable, this masks a critical failure mode: scanner variability systematically alters the embedding space and impacts calibration of downstream model predictions, resulting in scanner-dependent bias that can impact reliability in clinical use cases. We further show that robustness is not a simple function of training data scale, model size, or model recency. None of the models provided reliable robustness against scanner-induced variability. While the models trained on the most diverse data, here represented by vision-language models, appear to have an advantage with respect to robustness, they underperformed on downstream supervised tasks. We conclude that development and evaluation of PFMs requires moving beyond accuracy-centric benchmarks toward explicit evaluation and optimisation of embedding stability and calibration under realistic acquisition variability.
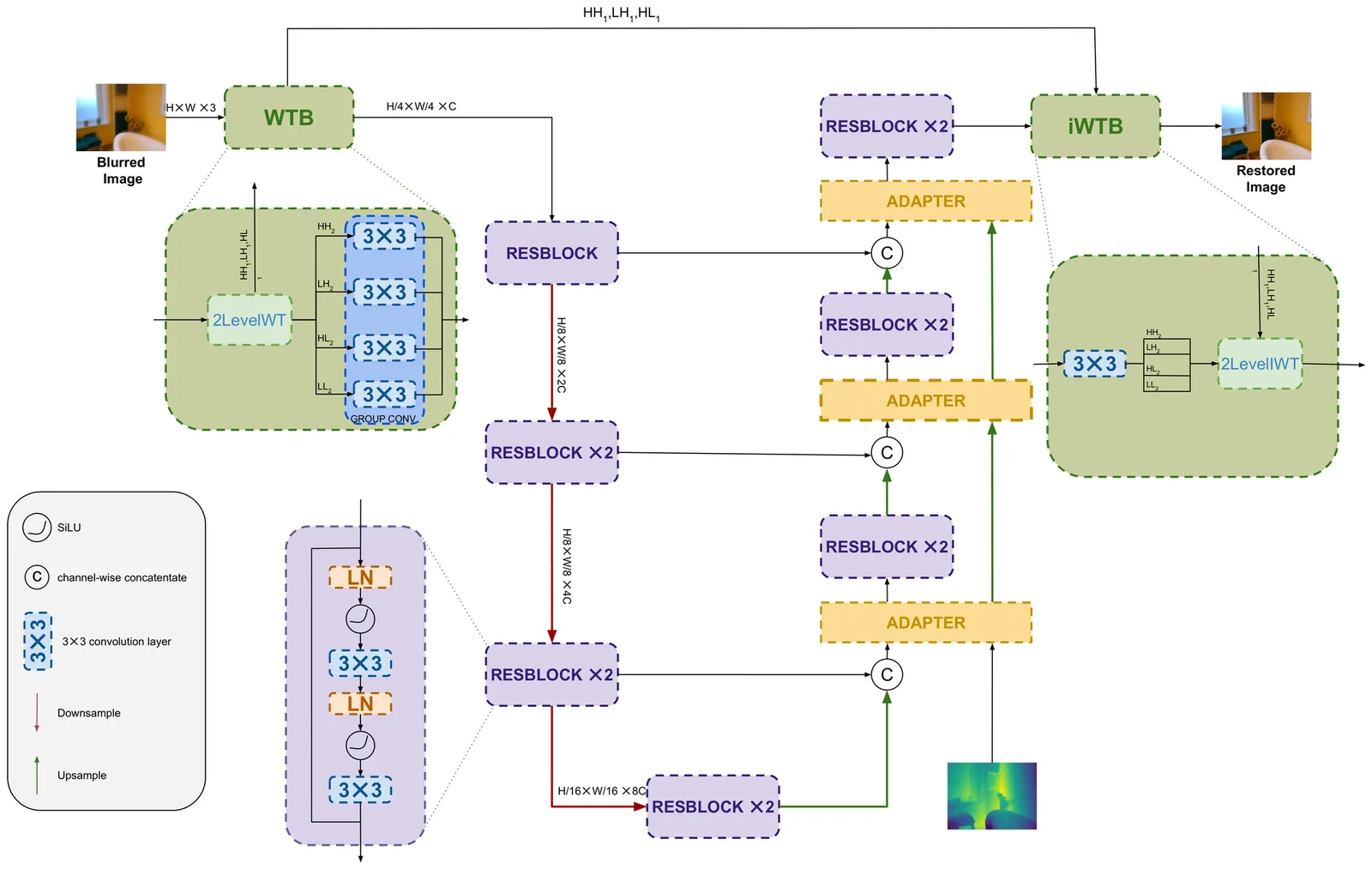
Image deblurring is a challenging problem in imaging due to its highly ill-posed nature. Deep learning models have shown great success in tackling this problem but the quest for the best image quality has brought their computational complexity up, making them impractical on anything but powerful servers. Meanwhile, recent works have shown that mobile Lidars can provide complementary information in the form of depth maps that enhance deblurring quality. In this paper, we introduce a novel low-complexity neural network for depth-guided image deblurring. We show that the use of the wavelet transform to separate structural details and reduce spatial redundancy as well as efficient feature conditioning on the depth information are essential ingredients in developing a low-complexity model. Experimental results show competitive image quality against recent state-of-the-art models while reducing complexity by up to two orders of magnitude.
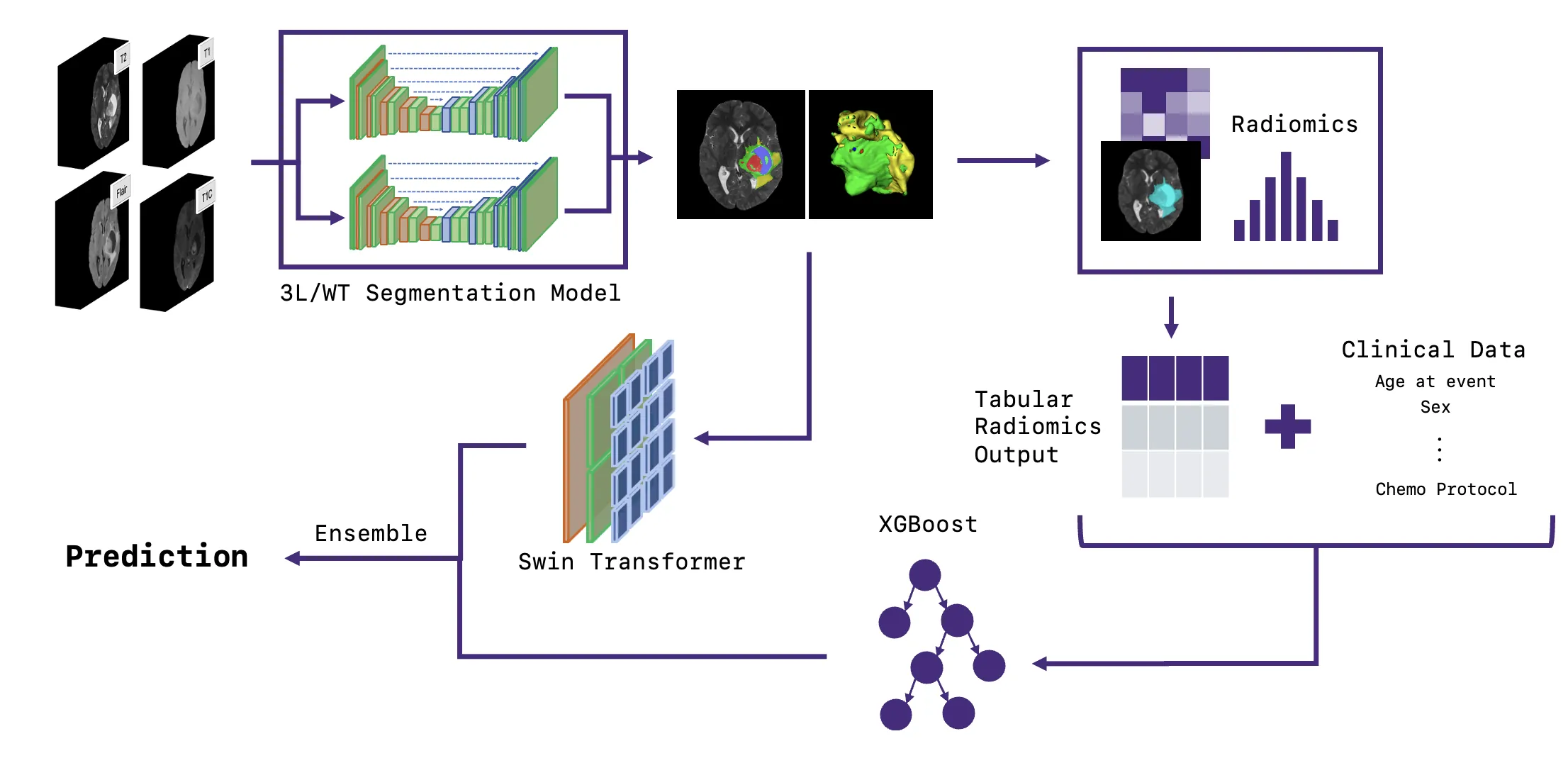
In this paper, we introduce a novel pipeline for predicting chemotherapy response in pediatric brain tumors that are not amenable to complete surgical resection, using pre-treatment magnetic resonance imaging combined with clinical information. Our method integrates a state-of-the-art pediatric brain tumor segmentation framework with radiomic feature extraction and clinical data through an ensemble of a Swin UNETR encoder and XGBoost classifier. The segmentation model delineates four tumor subregions enhancing tumor, non-enhancing tumor, cystic component and edema which are used to extract imaging biomarkers and generate predictive features. The Swin UNETR network classifies the response to treatment directly from these segmented MRI scans, while XGBoost predicts response using radiomics and clinical variables including legal sex, ethnicity, race, age at event (in days), molecular subtype, tumor locations, initial surgery status, metastatic status, metastasis location, chemotherapy type, protocol name and chemotherapy agents. The ensemble output provides a non-invasive estimate of chemotherapy response in this historically challenging population characterized by lower progression-free survival. Among compared approaches, our Swin-Ensemble achieved the best performance (precision for non effective cases=0.68, recall for non effective cases=0.85, precision for chemotherapy effective cases=0.64 and overall accuracy=0.69), outperforming Mamba-FeatureFuse, Swin UNETR encoder, and Swin-FeatureFuse models. Our findings suggest that this ensemble framework represents a promising step toward personalized therapy response prediction for pediatric low-grade glioma patients in need of chemotherapy treatment who are not suitable for complete surgical resection, a population with significantly lower progression free survival and for whom chemotherapy remains the primary treatment option.
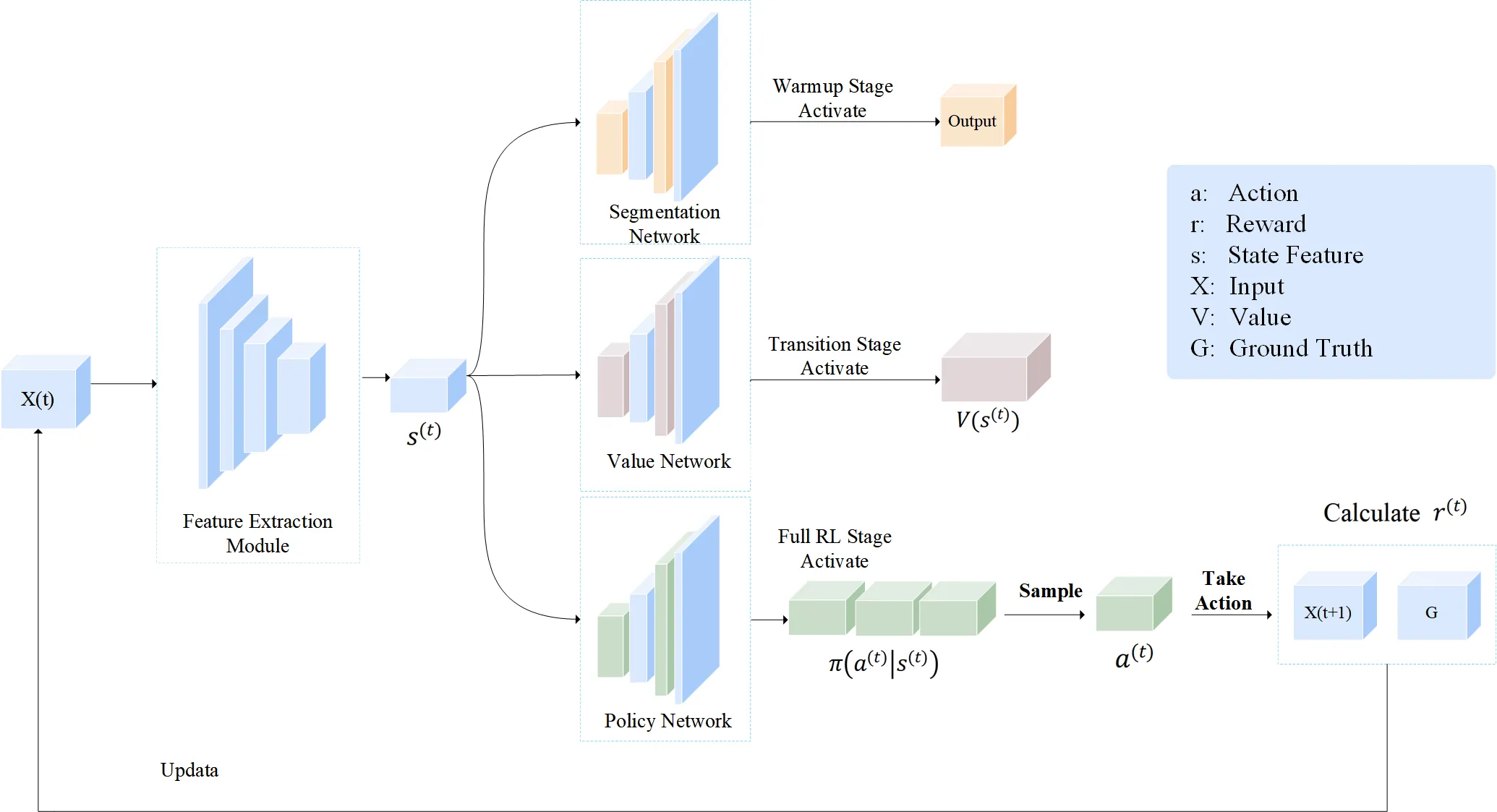
Deep learning has achieved significant advancements in medical image segmentation. Currently, obtaining accurate segmentation outcomes is critically reliant on large-scale datasets with high-quality annotations. However, noisy annotations are frequently encountered owing to the complex morphological structures of organs in medical images and variations among different annotators, which can substantially limit the efficacy of segmentation models. Motivated by the fact that medical imaging annotator can correct labeling errors during segmentation based on prior knowledge, we propose an end-to-end Staged Voxel-Level Deep Reinforcement Learning (SVL-DRL) framework for robust medical image segmentation under noisy annotations. This framework employs a dynamic iterative update strategy to automatically mitigate the impact of erroneous labels without requiring manual intervention. The key advancements of SVL-DRL over existing works include: i) formulating noisy annotations as a voxel-dependent problem and addressing it through a novel staged reinforcement learning framework which guarantees robust model convergence; ii) incorporating a voxel-level asynchronous advantage actor-critic (vA3C) module that conceptualizes each voxel as an autonomous agent, which allows each agent to dynamically refine its own state representation during training, thereby directly mitigating the influence of erroneous labels; iii) designing a novel action space for the agents, along with a composite reward function that strategically combines the Dice value and a spatial continuity metric to significantly boost segmentation accuracy while maintain semantic integrity. Experiments on three public medical image datasets demonstrates State-of-The-Art (SoTA) performance under various experimental settings, with an average improvement of over 3\% in both Dice and IoU scores.
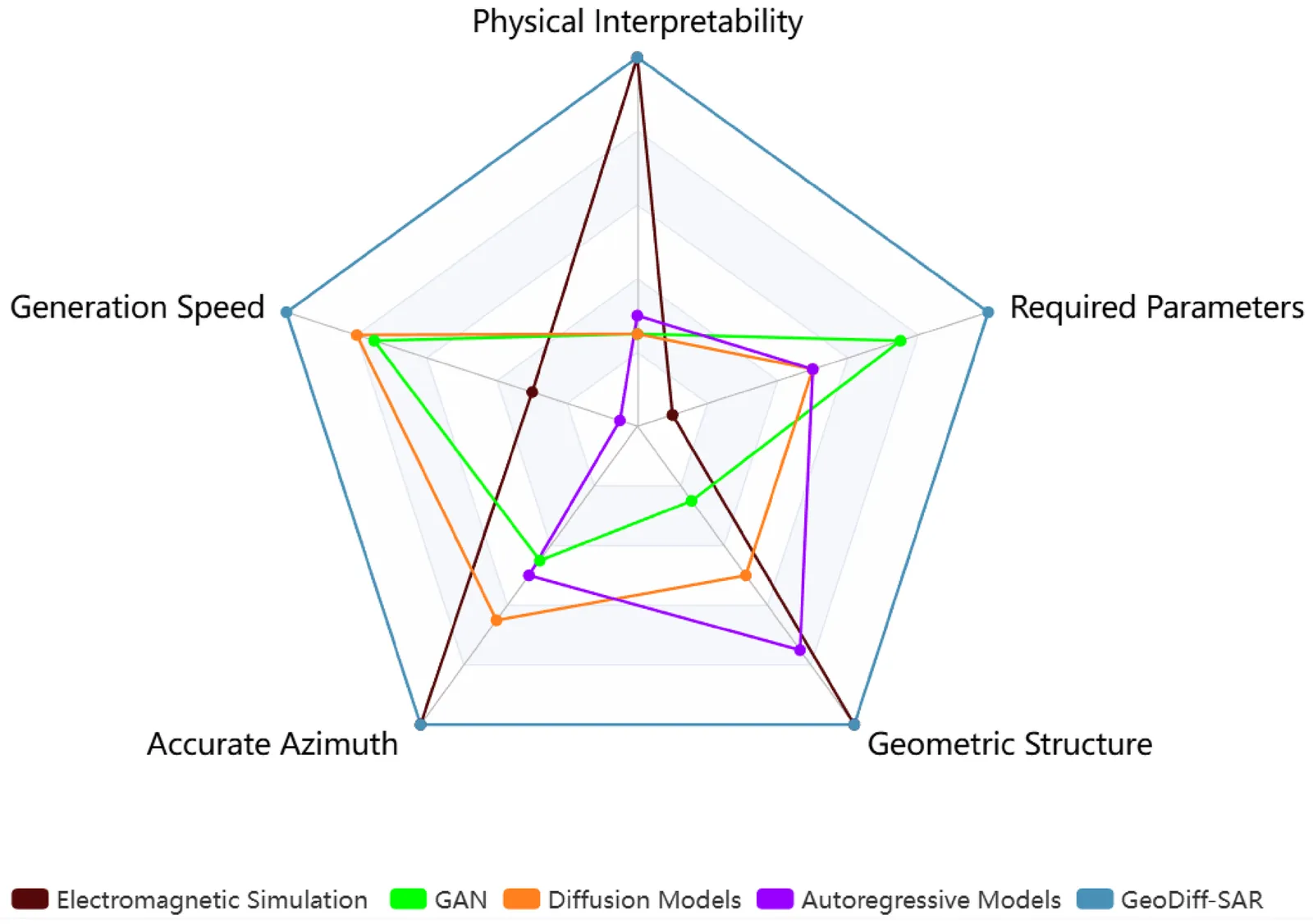
Synthetic Aperture Radar (SAR) imaging results are highly sensitive to observation geometries and the geometric parameters of targets. However, existing generative methods primarily operate within the image domain, neglecting explicit geometric information. This limitation often leads to unsatisfactory generation quality and the inability to precisely control critical parameters such as azimuth angles. To address these challenges, we propose GeoDiff-SAR, a geometric prior guided diffusion model for high-fidelity SAR image generation. Specifically, GeoDiff-SAR first efficiently simulates the geometric structures and scattering relationships inherent in real SAR imaging by calculating SAR point clouds at specific azimuths, which serves as a robust physical guidance. Secondly, to effectively fuse multi-modal information, we employ a feature fusion gating network based on Feature-wise Linear Modulation (FiLM) to dynamically regulate the weight distribution of 3D physical information, image control parameters, and textual description parameters. Thirdly, we utilize the Low-Rank Adaptation (LoRA) architecture to perform lightweight fine-tuning on the advanced Stable Diffusion 3.5 (SD3.5) model, enabling it to rapidly adapt to the distribution characteristics of the SAR domain. To validate the effectiveness of GeoDiff-SAR, extensive comparative experiments were conducted on real-world SAR datasets. The results demonstrate that data generated by GeoDiff-SAR exhibits high fidelity and effectively enhances the accuracy of downstream classification tasks. In particular, it significantly improves recognition performance across different azimuth angles, thereby underscoring the superiority of physics-guided generation.
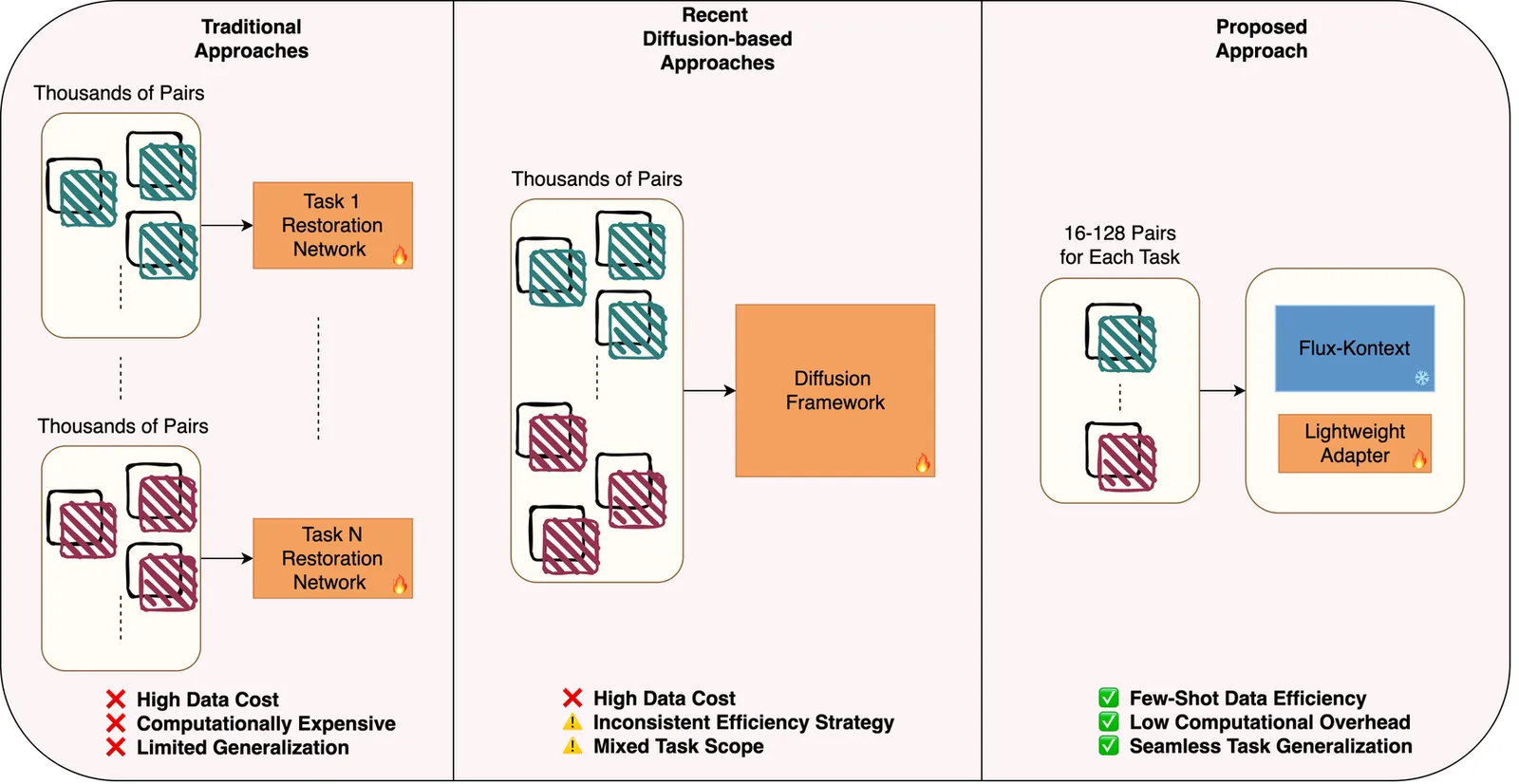
Image restoration has traditionally required training specialized models on thousands of paired examples per degradation type. We challenge this paradigm by demonstrating that powerful pre-trained text-conditioned image editing models can be efficiently adapted for multiple restoration tasks through parameter-efficient fine-tuning with remarkably few examples. Our approach fine-tunes LoRA adapters on FLUX.1 Kontext, a state-of-the-art 12B parameter flow matching model for image-to-image translation, using only 16-128 paired images per task, guided by simple text prompts that specify the restoration operation. Unlike existing methods that train specialized restoration networks from scratch with thousands of samples, we leverage the rich visual priors already encoded in large-scale pre-trained editing models, dramatically reducing data requirements while maintaining high perceptual quality. A single unified LoRA adapter, conditioned on task-specific text prompts, effectively handles multiple degradations including denoising, deraining, and dehazing. Through comprehensive ablation studies, we analyze: (i) the impact of training set size on restoration quality, (ii) trade-offs between task-specific versus unified multi-task adapters, (iii) the role of text encoder fine-tuning, and (iv) zero-shot baseline performance. While our method prioritizes perceptual quality over pixel-perfect reconstruction metrics like PSNR/SSIM, our results demonstrate that pre-trained image editing models, when properly adapted, offer a compelling and data-efficient alternative to traditional image restoration approaches, opening new avenues for few-shot, prompt-guided image enhancement. The code to reproduce our results are available at: https://github.com/makinyilmaz/Edit2Restore
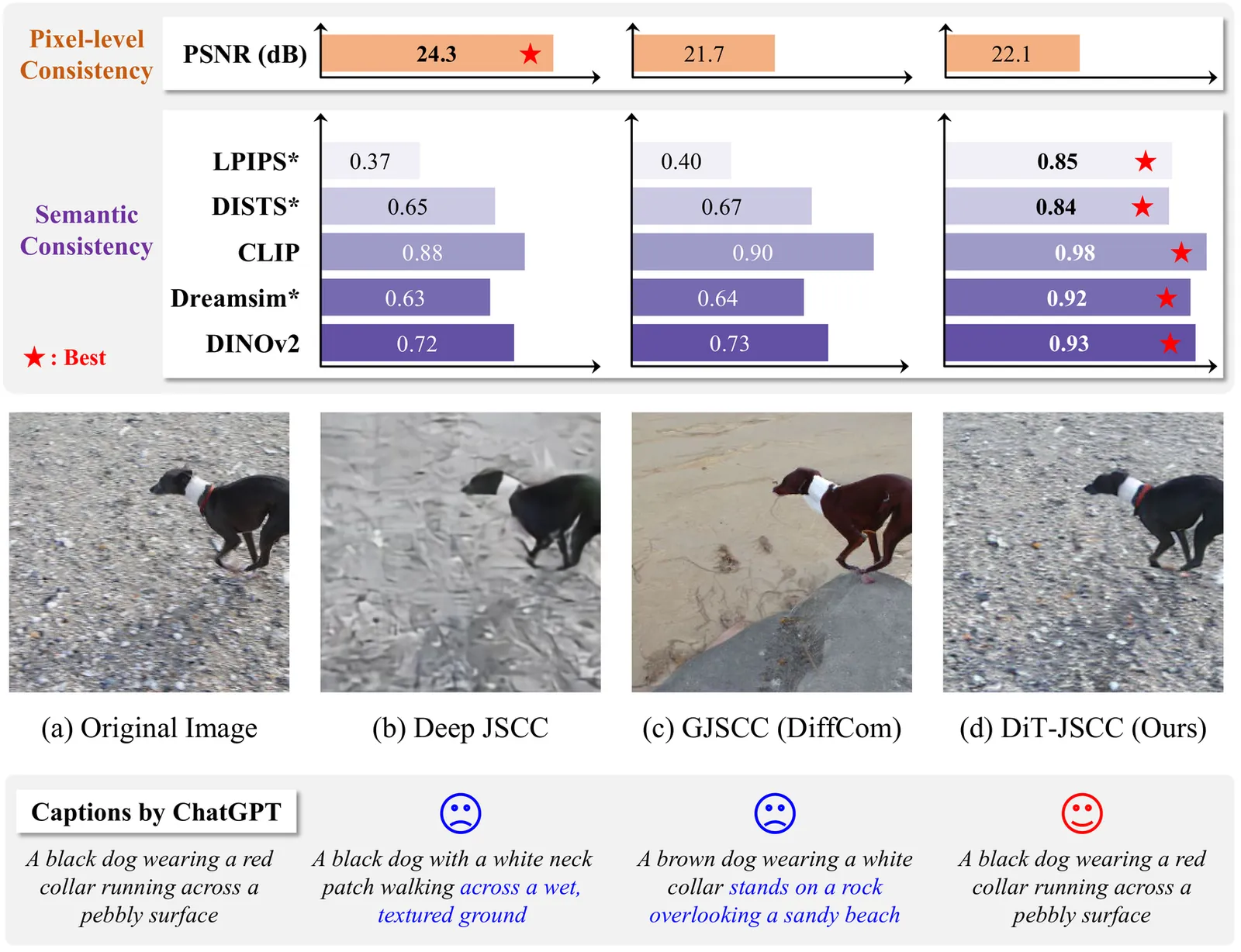
Generative joint source-channel coding (GJSCC) has emerged as a new Deep JSCC paradigm for achieving high-fidelity and robust image transmission under extreme wireless channel conditions, such as ultra-low bandwidth and low signal-to-noise ratio. Recent studies commonly adopt diffusion models as generative decoders, but they frequently produce visually realistic results with limited semantic consistency. This limitation stems from a fundamental mismatch between reconstruction-oriented JSCC encoders and generative decoders, as the former lack explicit semantic discriminability and fail to provide reliable conditional cues. In this paper, we propose DiT-JSCC, a novel GJSCC backbone that can jointly learn a semantics-prioritized representation encoder and a diffusion transformer (DiT) based generative decoder, our open-source project aims to promote the future research in GJSCC. Specifically, we design a semantics-detail dual-branch encoder that aligns naturally with a coarse-to-fine conditional DiT decoder, prioritizing semantic consistency under extreme channel conditions. Moreover, a training-free adaptive bandwidth allocation strategy inspired by Kolmogorov complexity is introduced to further improve the transmission efficiency, thereby indeed redefining the notion of information value in the era of generative decoding. Extensive experiments demonstrate that DiT-JSCC consistently outperforms existing JSCC methods in both semantic consistency and visual quality, particularly in extreme regimes.
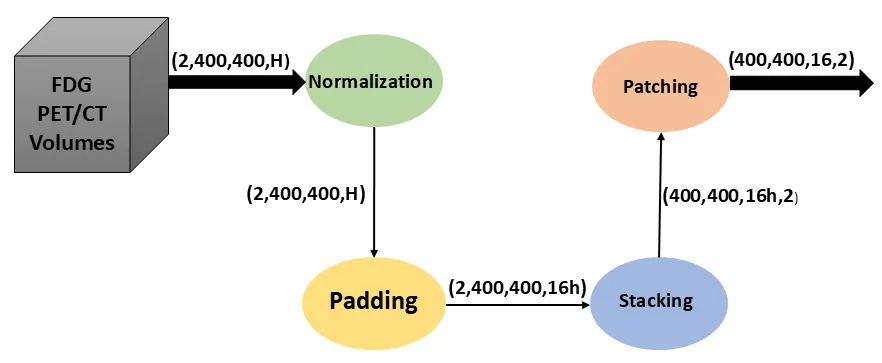
Accurate and automated lesion segmentation in Positron Emission Tomography / Computed Tomography (PET/CT) imaging is essential for cancer diagnosis and therapy planning. This paper presents a Swin Transformer UNet 3D (SwinUNet3D) framework for lesion segmentation in Fluorodeoxyglucose Positron Emission Tomography / Computed Tomography (FDG-PET/CT) scans. By combining shifted window self-attention with U-Net style skip connections, the model captures both global context and fine anatomical detail. We evaluate SwinUNet3D on the AutoPET III FDG dataset and compare it against a baseline 3D U-Net. Results show that SwinUNet3D achieves a Dice score of 0.88 and IoU of 0.78, surpassing 3D U-Net (Dice 0.48, IoU 0.32) while also delivering faster inference times. Qualitative analysis demonstrates improved detection of small and irregular lesions, reduced false positives, and more accurate PET/CT fusion. While the framework is currently limited to FDG scans and trained under modest GPU resources, it establishes a strong foundation for future multi-tracer, multi-center evaluations and benchmarking against other transformer-based architectures. Overall, SwinUNet3D represents an efficient and robust approach to PET/CT lesion segmentation, advancing the integration of transformer-based models into oncology imaging workflows.
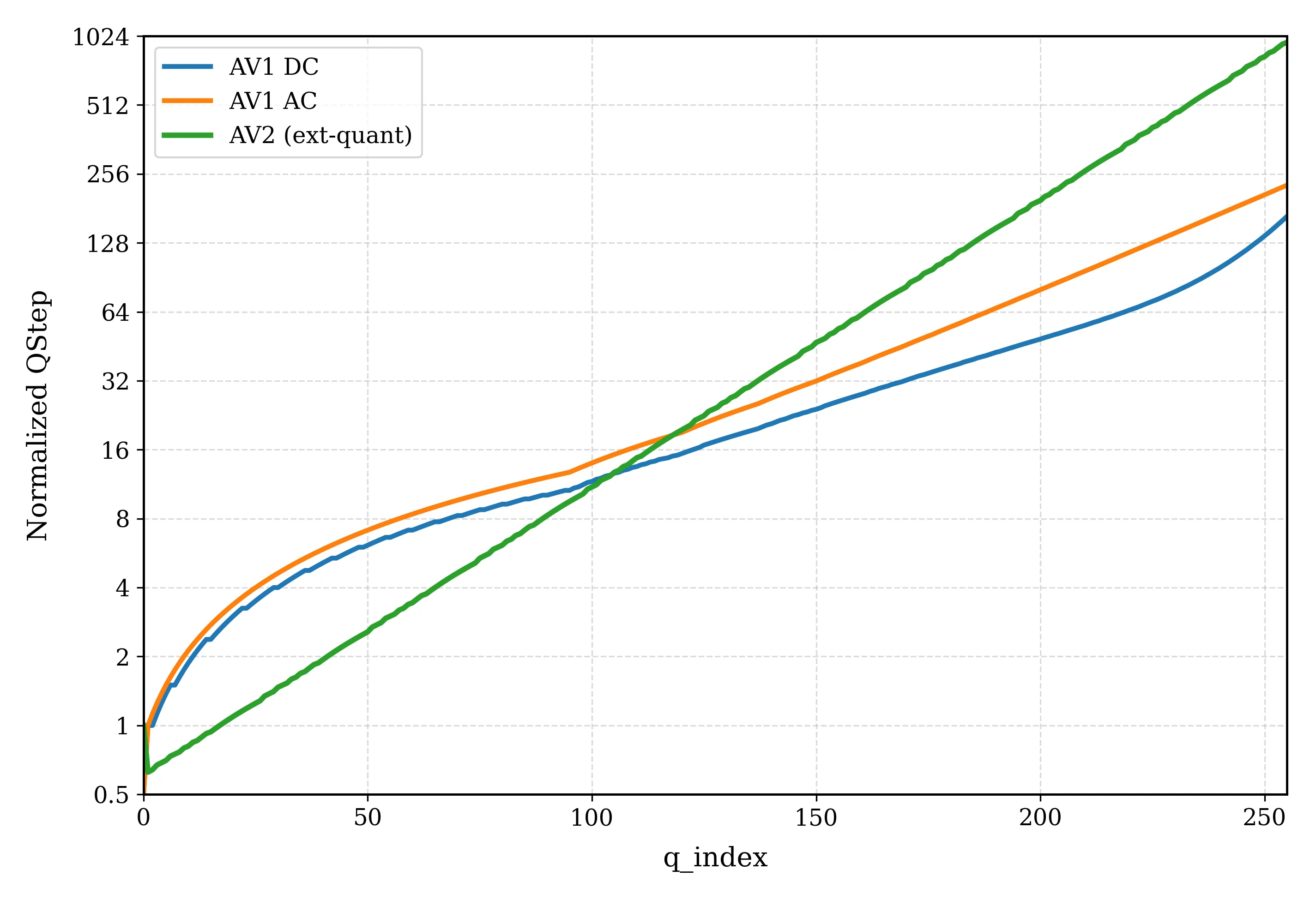
AV2 is the successor to the AV1 royalty-free video coding standard developed by the Alliance for Open Media (AOMedia). Its primary objective is to deliver substantial compression gains and subjective quality improvements while maintaining low-complexity encoder and decoder operations. This paper describes the transform, quantization and entropy coding design in AV2, including redesigned transform kernels and data-driven transforms, expanded transform partitioning, and a mode & coefficient dependent transform signaling. AV2 introduces several new coding tools including Intra/Inter Secondary Transforms (IST), Trellis Coded Quantization (TCQ), Adaptive Transform Coding (ATC), Probability Adaptation Rate Adjustment (PARA), Forward Skip Coding (FSC), Cross Chroma Component Transforms (CCTX), Parity Hiding (PH) tools and improved lossless coding. These advances enable AV2 to deliver the highest quality video experience for video applications at a significantly reduced bitrate.

Solving inverse problems in imaging requires models that support efficient inference, uncertainty quantification, and principled probabilistic reasoning. Energy-Based Models (EBMs), with their interpretable energy landscapes and compositional structure, are well-suited for this task but have historically suffered from high computational costs and training instability. To overcome the historical shortcomings of EBMs, we introduce a fast distillation strategy to transfer the strengths of pre-trained diffusion models into multi-scale EBMs. These distilled EBMs enable efficient sampling and preserve the interpretability and compositionality inherent to potential-based frameworks. Leveraging EBM compositionality, we propose Annealed Langevin Posterior Sampling (ALPS) algorithm for Maximum-A-Posteriori (MAP), Minimum Mean Square Error (MMSE), and uncertainty estimates for inverse problems in imaging. Unlike diffusion models that use complex guidance strategies for latent variables, we perform annealing on static posterior distributions that are well-defined and composable. Experiments on image inpainting and MRI reconstruction demonstrate that our method matches or surpasses diffusion-based baselines in both accuracy and efficiency, while also supporting MAP recovery. Overall, our framework offers a scalable and principled solution for inverse problems in imaging, with potential for practical deployment in scientific and clinical settings. ALPS code is available at the GitHub repository \href{https://github.com/JyoChand/ALPS}{ALPS}.
Accurate crop yield prediction relies on diverse data streams, including satellite, meteorological, soil, and topographic information. However, despite rapid advances in machine learning, existing approaches remain crop- or region-specific and require data engineering efforts. This limits scalability, reproducibility, and operational deployment. This study introduces UniCrop, a universal and reusable data pipeline designed to automate the acquisition, cleaning, harmonisation, and engineering of multi-source environmental data for crop yield prediction. For any given location, crop type, and temporal window, UniCrop automatically retrieves, harmonises, and engineers over 200 environmental variables (Sentinel-1/2, MODIS, ERA5-Land, NASA POWER, SoilGrids, and SRTM), reducing them to a compact, analysis-ready feature set utilising a structured feature reduction workflow with minimum redundancy maximum relevance (mRMR). To validate, UniCrop was applied to a rice yield dataset comprising 557 field observations. Using only the selected 15 features, four baseline machine learning models (LightGBM, Random Forest, Support Vector Regression, and Elastic Net) were trained. LightGBM achieved the best single-model performance (RMSE = 465.1 kg/ha, $R^2 = 0.6576$), while a constrained ensemble of all baselines further improved accuracy (RMSE = 463.2 kg/ha, $R^2 = 0.6604$). UniCrop contributes a scalable and transparent data-engineering framework that addresses the primary bottleneck in operational crop yield modelling: the preparation of consistent and harmonised multi-source data. By decoupling data specification from implementation and supporting any crop, region, and time frame through simple configuration updates, UniCrop provides a practical foundation for scalable agricultural analytics. The code and implementation documentation are shared in https://github.com/CoDIS-Lab/UniCrop.

Synthetic aperture radar (SAR) provides valuable information about the Earth's surface under all weather and illumination conditions. However, the inherent phenomenon of speckle and the presence of sidelobes around bright targets pose challenges for accurate interpretation of SAR imagery. Most existing SAR image restoration methods address despeckling and sidelobes reduction as separate tasks. In this paper, we propose a unified framework that jointly performs both tasks using neural networks (NNs) trained on a realistic SAR simulated dataset generated with MOCEM. Inference can then be performed on real SAR images, demonstrating effective simulation to real (Sim2Real) transferability. Additionally, we incorporate acquisition metadata as auxiliary input to the NNs, demonstrating improved restoration performance.
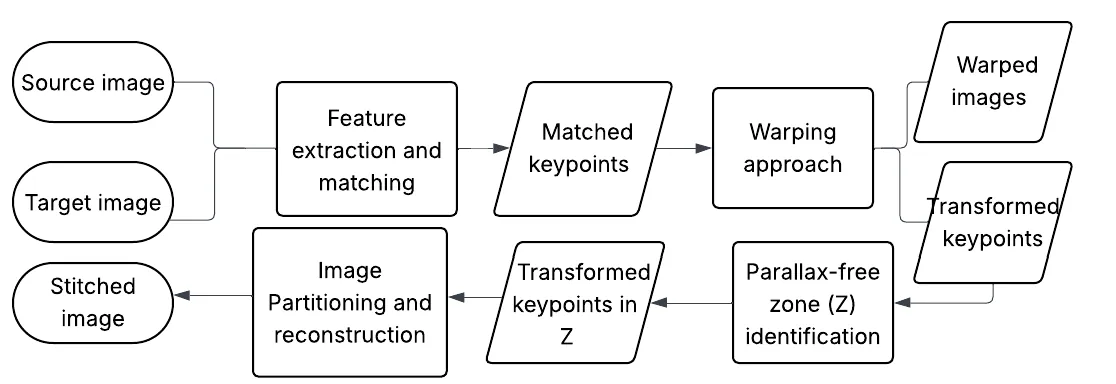
This paper introduces SENA (SEamlessly NAtural), a geometry-driven image stitching approach that prioritizes structural fidelity in challenging real-world scenes characterized by parallax and depth variation. Conventional image stitching relies on homographic alignment, but this rigid planar assumption often fails in dual-camera setups with significant scene depth, leading to distortions such as visible warps and spherical bulging. SENA addresses these fundamental limitations through three key contributions. First, we propose a hierarchical affine-based warping strategy, combining global affine initialization with local affine refinement and smooth free-form deformation. This design preserves local shape, parallelism, and aspect ratios, thereby avoiding the hallucinated structural distortions commonly introduced by homography-based models. Second, we introduce a geometry-driven adequate zone detection mechanism that identifies parallax-minimized regions directly from the disparity consistency of RANSAC-filtered feature correspondences, without relying on semantic segmentation. Third, building upon this adequate zone, we perform anchor-based seamline cutting and segmentation, enforcing a one-to-one geometric correspondence across image pairs by construction, which effectively eliminates ghosting, duplication, and smearing artifacts in the final panorama. Extensive experiments conducted on challenging datasets demonstrate that SENA achieves alignment accuracy comparable to leading homography-based methods, while significantly outperforming them in critical visual metrics such as shape preservation, texture integrity, and overall visual realism.
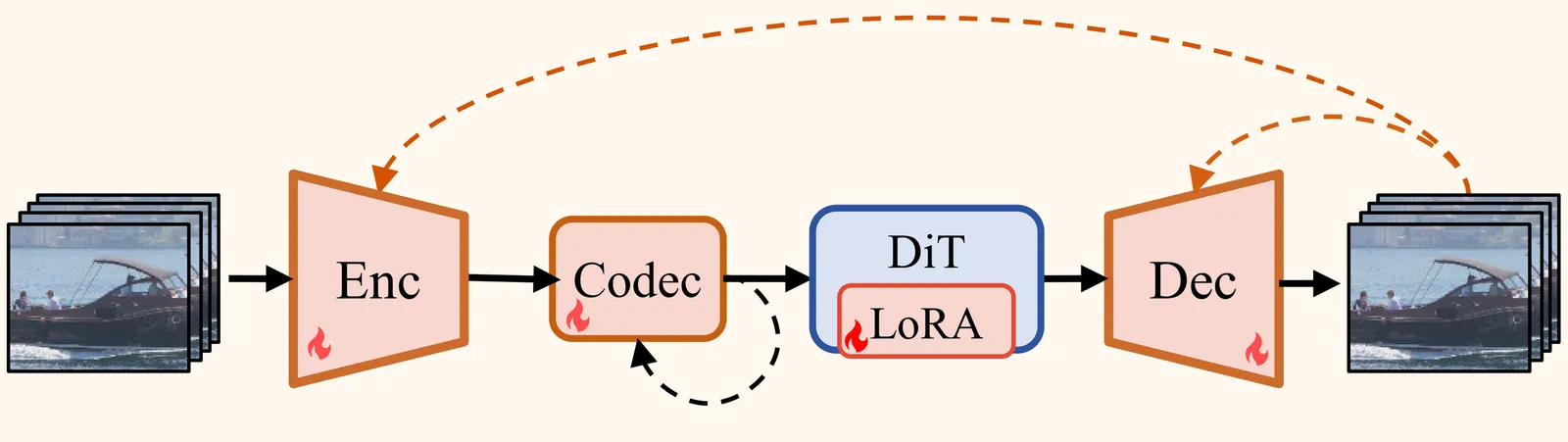
While one-step diffusion models have recently excelled in perceptual image compression, their application to video remains limited. Prior efforts typically rely on pretrained 2D autoencoders that generate per-frame latent representations independently, thereby neglecting temporal dependencies. We present YODA--Yet Another One-step Diffusion-based Video Compressor--which embeds multiscale features from temporal references for both latent generation and latent coding to better exploit spatial-temporal correlations for more compact representation, and employs a linear Diffusion Transformer (DiT) for efficient one-step denoising. YODA achieves state-of-the-art perceptual performance, consistently outperforming traditional and deep-learning baselines on LPIPS, DISTS, FID, and KID. Source code will be publicly available at https://github.com/NJUVISION/YODA.
Artificial intelligence models have shown strong potential in acute ischemic stroke imaging, particularly for lesion detection and segmentation using computed tomography and magnetic resonance imaging. However, most existing approaches operate as black box predictors, producing deterministic outputs without explicit uncertainty awareness or structured mechanisms to abstain under ambiguous conditions. This limitation raises serious safety and trust concerns in high risk emergency radiology settings. In this paper, we propose an explainable agentic AI framework for uncertainty aware and abstention enabled decision support in acute ischemic stroke imaging. The framework follows a modular agentic pipeline in which a perception agent performs lesion aware image analysis, an uncertainty estimation agent computes slice level predictive reliability, and a decision agent determines whether to issue a prediction or abstain based on predefined uncertainty thresholds. Unlike prior stroke imaging systems that primarily focus on improving segmentation or classification accuracy, the proposed framework explicitly prioritizes clinical safety, transparency, and clinician aligned decision behavior. Qualitative and case based analyses across representative stroke imaging scenarios demonstrate that uncertainty driven abstention naturally emerges in diagnostically ambiguous regions and low information slices. The framework further integrates visual explanation mechanisms to support both predictive and abstention decisions, addressing a key limitation of existing uncertainty aware medical imaging systems. Rather than introducing a new performance benchmark, this work presents agentic control, uncertainty awareness, and selective abstention as essential design principles for developing safe and trustworthy medical imaging AI systems.
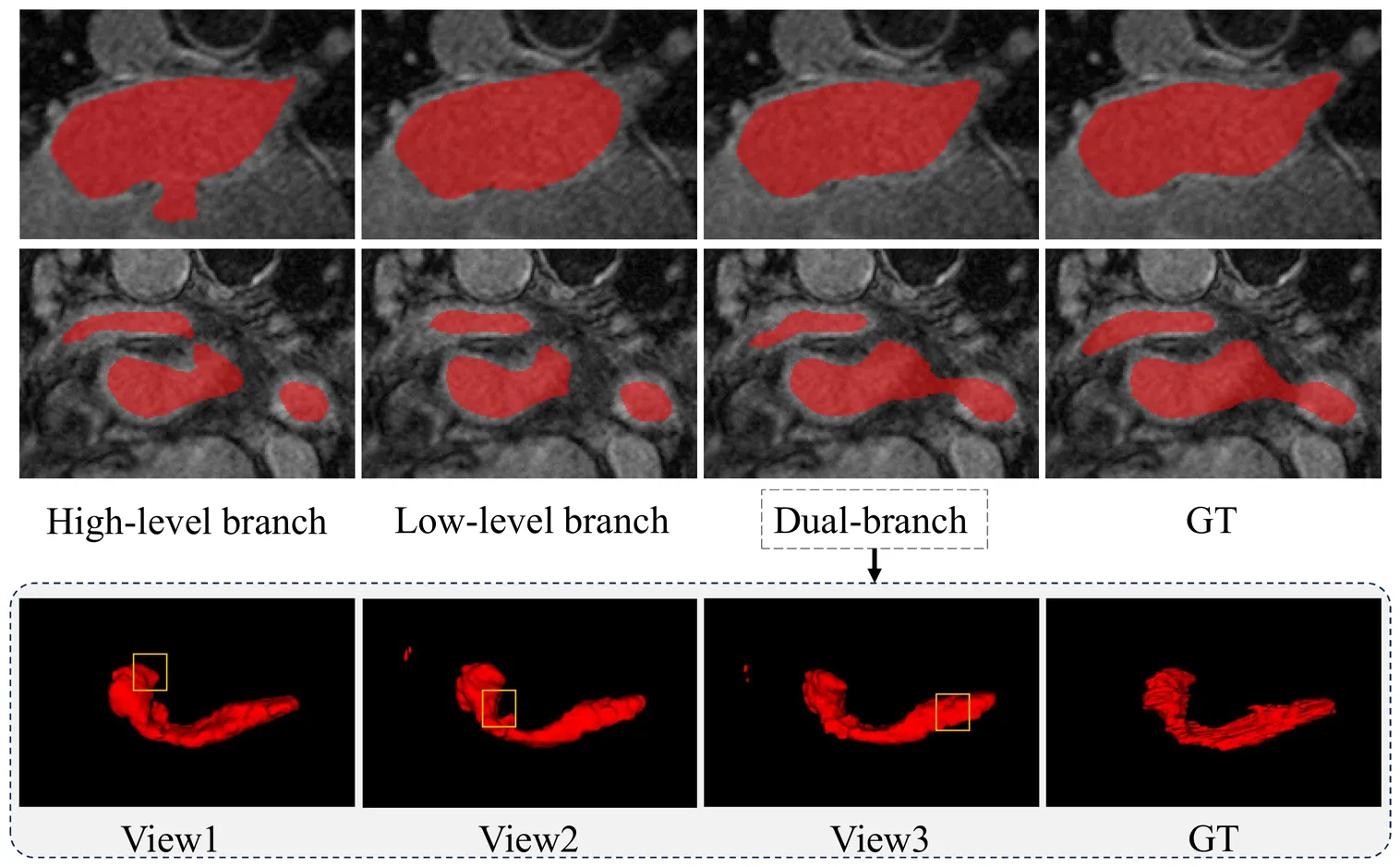
Medical image segmentation faces critical challenges in semi-supervised learning scenarios due to severe annotation scarcity requiring expert radiological knowledge, significant inter-annotator variability across different viewpoints and expertise levels, and inadequate multi-scale feature integration for precise boundary delineation in complex anatomical structures. Existing semi-supervised methods demonstrate substantial performance degradation compared to fully supervised approaches, particularly in small target segmentation and boundary refinement tasks. To address these fundamental challenges, we propose SASNet (Scale-aware Adaptive Supervised Network), a dual-branch architecture that leverages both low-level and high-level feature representations through novel scale-aware adaptive reweight mechanisms. Our approach introduces three key methodological innovations, including the Scale-aware Adaptive Reweight strategy that dynamically weights pixel-wise predictions using temporal confidence accumulation, the View Variance Enhancement mechanism employing 3D Fourier domain transformations to simulate annotation variability, and segmentation-regression consistency learning through signed distance map algorithms for enhanced boundary precision. These innovations collectively address the core limitations of existing semi-supervised approaches by integrating spatial, temporal, and geometric consistency principles within a unified optimization framework. Comprehensive evaluation across LA, Pancreas-CT, and BraTS datasets demonstrates that SASNet achieves superior performance with limited labeled data, surpassing state-of-the-art semi-supervised methods while approaching fully supervised performance levels. The source code for SASNet is available at https://github.com/HUANGLIZI/SASNet.
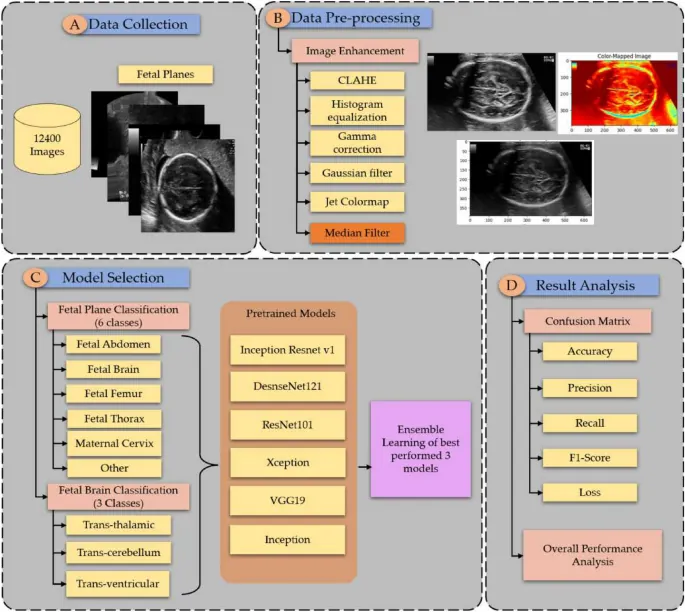
Fetal ultrasound standard-plane classification underpins reliable prenatal biometry and anomaly screening, yet real-world deployment is limited by domain shift, image noise, and poor calibration of predicted probabilities. This paper presents a practical framework for uncertainty-calibrated explainable AI in fetal plane classification. We synthesize uncertainty estimation methods (Monte Carlo dropout, deep ensembles, evidential learning, and conformal prediction) with post-hoc and uncertainty-aware explanations (Grad-CAM variants, LIME-style local surrogates, and uncertainty-weighted multi-resolution activation maps), and we map these components to a clinician-facing workflow. Using FETAL_PLANES_DB as a reference benchmark, we define a reporting protocol that couples accuracy with calibration and selective prediction, including expected calibration error, Brier score, coverage-risk curves, and structured error analysis with explanations. We also discuss integration points for quality control and human-in-the-loop review, where uncertainty flags trigger re-acquisition or expert confirmation. The goal is a reproducible, clinically aligned blueprint for building fetal ultrasound classifiers whose confidence and explanations remain trustworthy under noisy acquisition conditions.
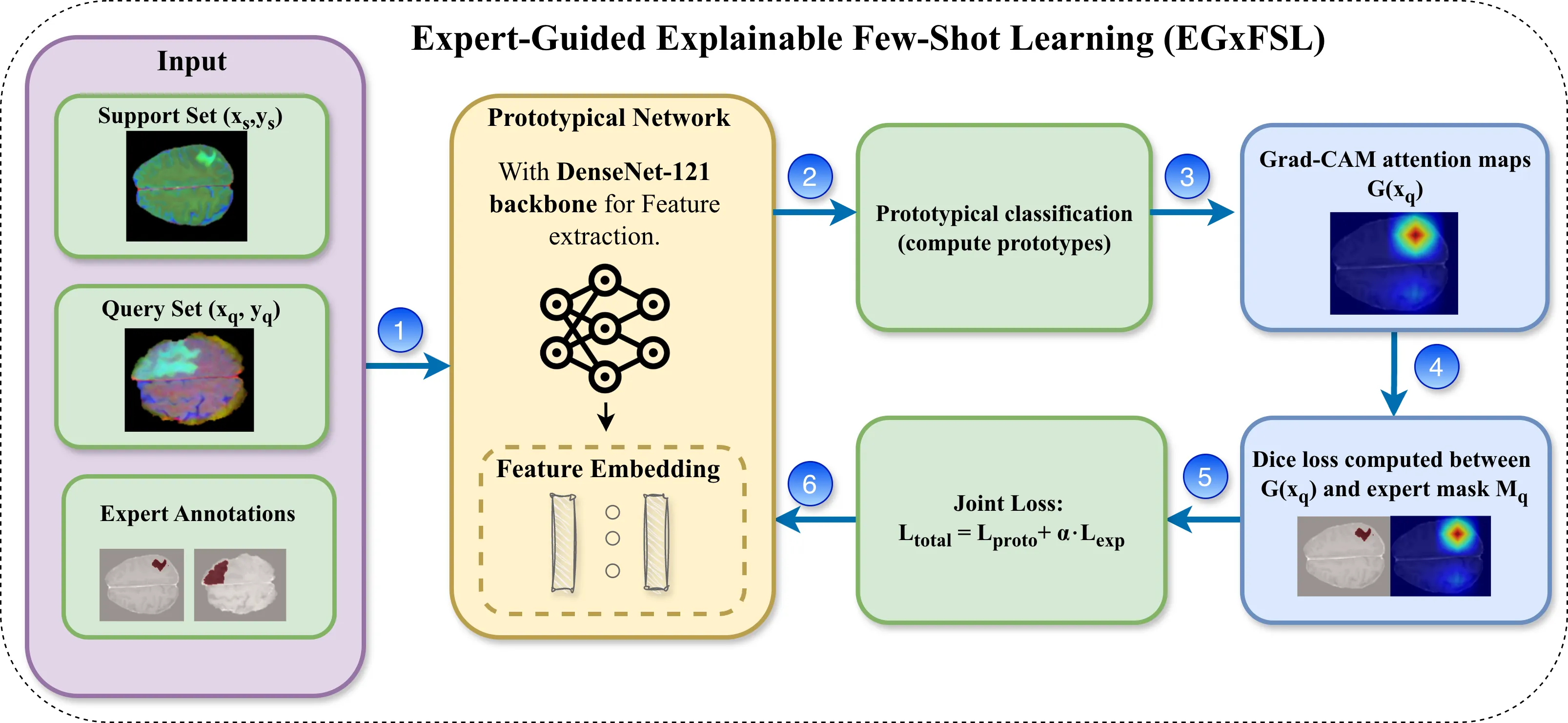
Medical image analysis faces two critical challenges: scarcity of labeled data and lack of model interpretability, both hindering clinical AI deployment. Few-shot learning (FSL) addresses data limitations but lacks transparency in predictions. Active learning (AL) methods optimize data acquisition but overlook interpretability of acquired samples. We propose a dual-framework solution: Expert-Guided Explainable Few-Shot Learning (EGxFSL) and Explainability-Guided AL (xGAL). EGxFSL integrates radiologist-defined regions-of-interest as spatial supervision via Grad-CAM-based Dice loss, jointly optimized with prototypical classification for interpretable few-shot learning. xGAL introduces iterative sample acquisition prioritizing both predictive uncertainty and attention misalignment, creating a closed-loop framework where explainability guides training and sample selection synergistically. On the BraTS (MRI), VinDr-CXR (chest X-ray), and SIIM-COVID-19 (chest X-ray) datasets, we achieve accuracies of 92\%, 76\%, and 62\%, respectively, consistently outperforming non-guided baselines across all datasets. Under severe data constraints, xGAL achieves 76\% accuracy with only 680 samples versus 57\% for random sampling. Grad-CAM visualizations demonstrate guided models focus on diagnostically relevant regions, with generalization validated on breast ultrasound confirming cross-modality applicability.
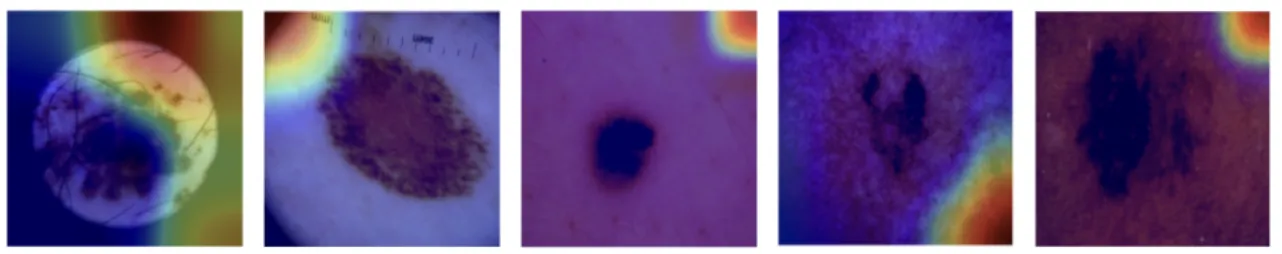
Melanoma is the most lethal subtype of skin cancer, and early and accurate detection of this disease can greatly improve patients' outcomes. Although machine learning models, especially convolutional neural networks (CNNs), have shown great potential in automating melanoma classification, their diagnostic reliability still suffers due to inconsistent focus on lesion areas. In this study, we analyze the relationship between lesion attention and diagnostic performance, involving masked images, bounding box detection, and transfer learning. We used multiple explainability and sensitivity analysis approaches to investigate how well models aligned their attention with lesion areas and how this alignment correlated with precision, recall, and F1-score. Results showed that models with a higher focus on lesion areas achieved better diagnostic performance, suggesting the potential of interpretable AI in medical diagnostics. This study provides a foundation for developing more accurate and trustworthy melanoma classification models in the future.
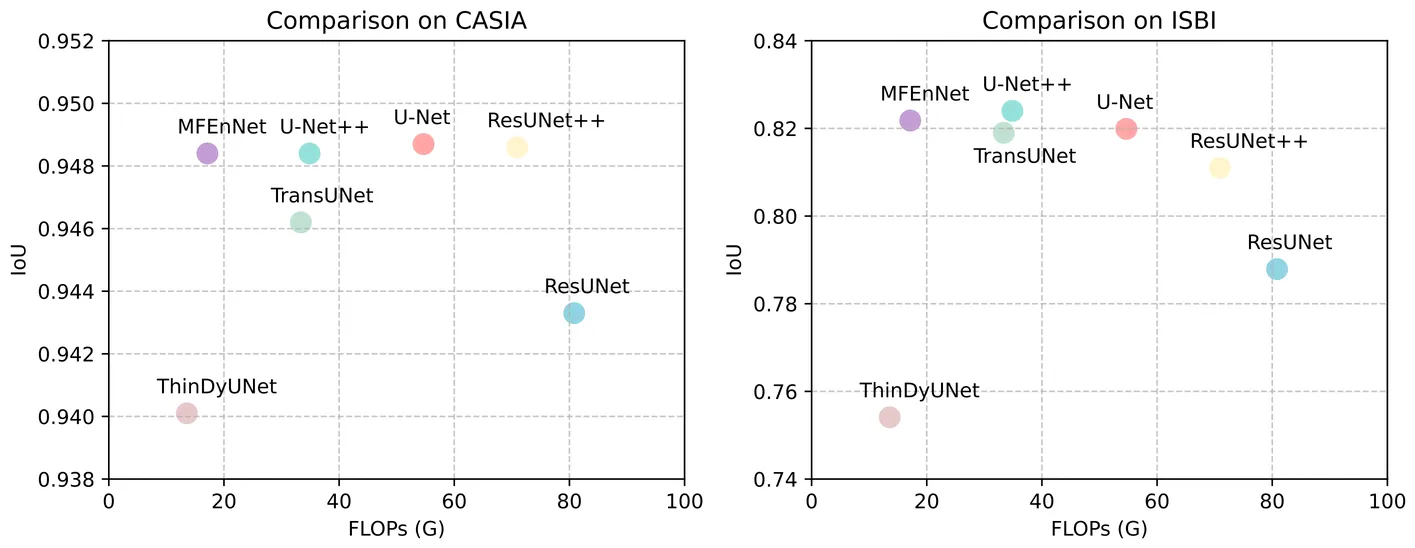
Semantic segmentation is crucial for medical image analysis, enabling precise disease diagnosis and treatment planning. However, many advanced models employ complex architectures, limiting their use in resource-constrained clinical settings. This paper proposes MFEnNet, an efficient medical image segmentation framework that incorporates MetaFormer in the encoding phase of the U-Net backbone. MetaFormer, an architectural abstraction of vision transformers, provides a versatile alternative to convolutional neural networks by transforming tokenized image patches into sequences for global context modeling. To mitigate the substantial computational cost associated with self-attention, the proposed framework replaces conventional transformer modules with pooling transformer blocks, thereby achieving effective global feature aggregation at reduced complexity. In addition, Swish activation is used to achieve smoother gradients and faster convergence, while spatial pyramid pooling is incorporated at the bottleneck to improve multi-scale feature extraction. Comprehensive experiments on different medical segmentation benchmarks demonstrate that the proposed MFEnNet approach attains competitive accuracy while significantly lowering computational cost compared to state-of-the-art models. The source code for this work is available at https://github.com/tranleanh/mfennet.