Biomolecules
Molecular biology, protein structure, and biochemistry
Molecular biology, protein structure, and biochemistry
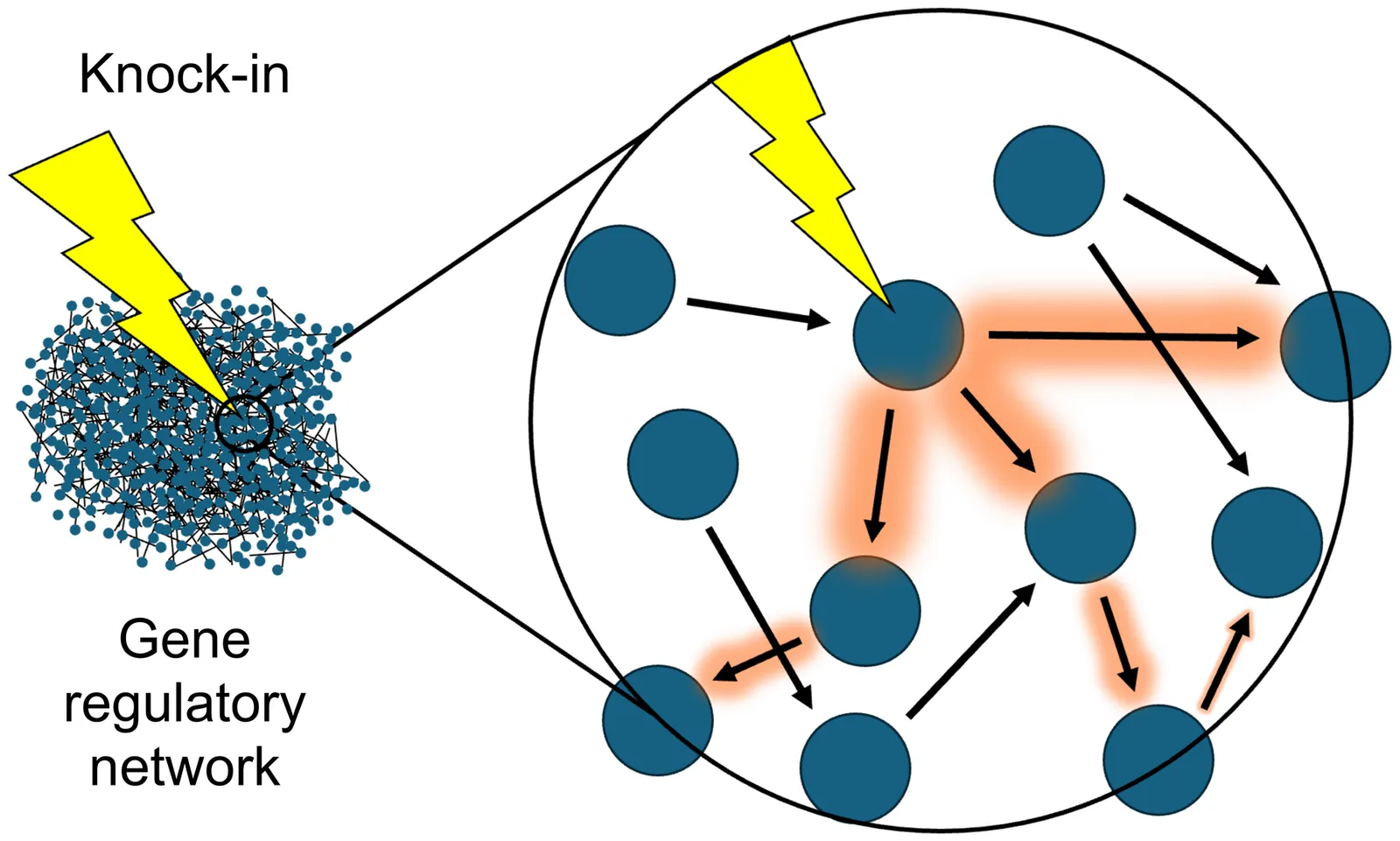
A hallmark of aging is loss of information in gene regulatory networks. These networks are tightly connected, raising the question of whether information could be restored by perturbing single genes. We develop a simple theoretical framework for information transmission in gene regulatory networks that describes the information gained or lost when a gene is "knocked in" (exogenously expressed). Applying the framework to gene expression data from muscle cells in young and old mice, we find that single knock-ins can restore network information by up to 10%. Our work advances the study of information flow in networks and identifies potential gene targets for rejuvenation.
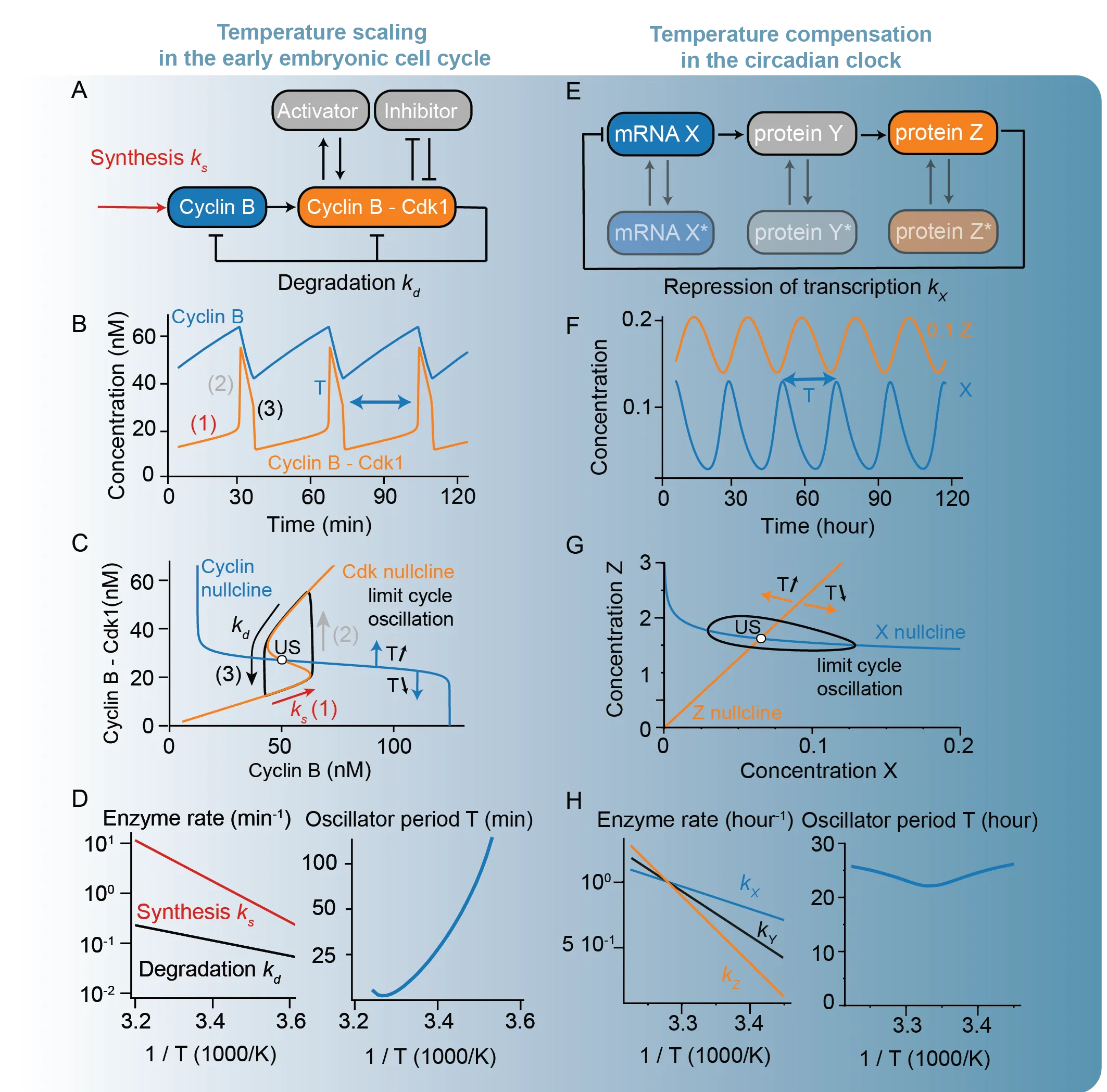
Building on the phenomenological and microscopic models reviewed in Part I, this second part focuses on network-level mechanisms that generate emergent temperature response curves. We review deterministic models in which temperature modulates the kinetics of coupled biochemical reactions, as well as stochastic frameworks, such as Markov chains, that capture more complex multi-step processes. These approaches show how Arrhenius-like temperature dependence at the level of individual reactions is transformed into non-Arrhenius scaling, thermal limits, and temperature compensation at the system level. Together, network-level models provide a mechanistic bridge between empirical temperature response curves and the molecular organization of biological systems, giving us predictive insights into robustness, perturbations, and evolutionary constraints.
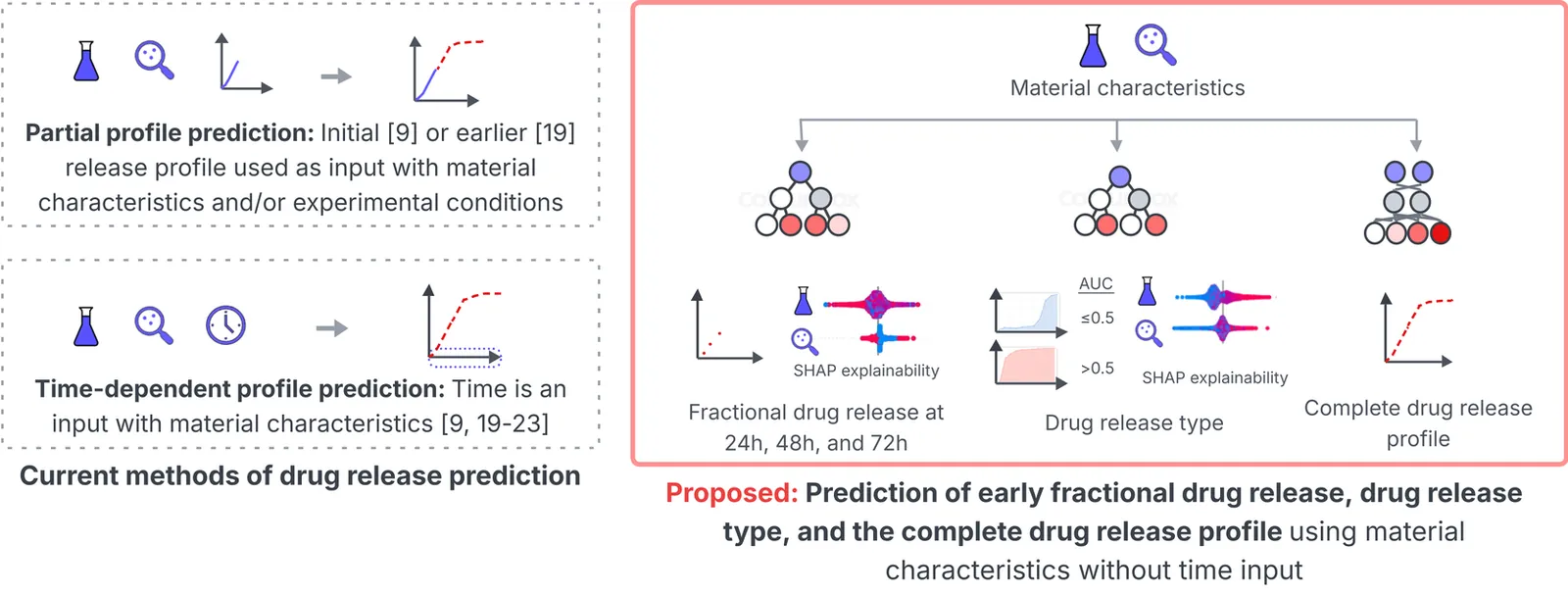
Polymer-based long-acting injectables (LAIs) have transformed the treatment of chronic diseases by enabling controlled drug delivery, thus reducing dosing frequency and extending therapeutic duration. Achieving controlled drug release from LAIs requires extensive optimization of the complex underlying physicochemical properties. Machine learning (ML) can accelerate LAI development by modeling the complex relationships between LAI properties and drug release. However, recent ML studies have provided limited information on key properties that modulate drug release, due to the lack of custom modeling and analysis tailored to LAI data. This paper presents a novel data transformation and explainable ML approach to synthesize actionable information from 321 LAI formulations by predicting early drug release at 24, 48, and 72 hours, classification of release profile types, and prediction of complete release profiles. These three experiments investigate the contribution and control of LAI material characteristics in early and complete drug release profiles. A strong correlation (>0.65) is observed between the true and predicted drug release in 72 hours, while a 0.87 F1-score is obtained in classifying release profile types. A time-independent ML framework predicts delayed biphasic and triphasic curves with better performance than current time-dependent approaches. Shapley additive explanations reveal the relative influence of material characteristics during early and for complete release which fill several gaps in previous in-vitro and ML-based studies. The novel approach and findings can provide a quantitative strategy and recommendations for scientists to optimize the drug-release dynamics of LAI. The source code for the model implementation is publicly available.
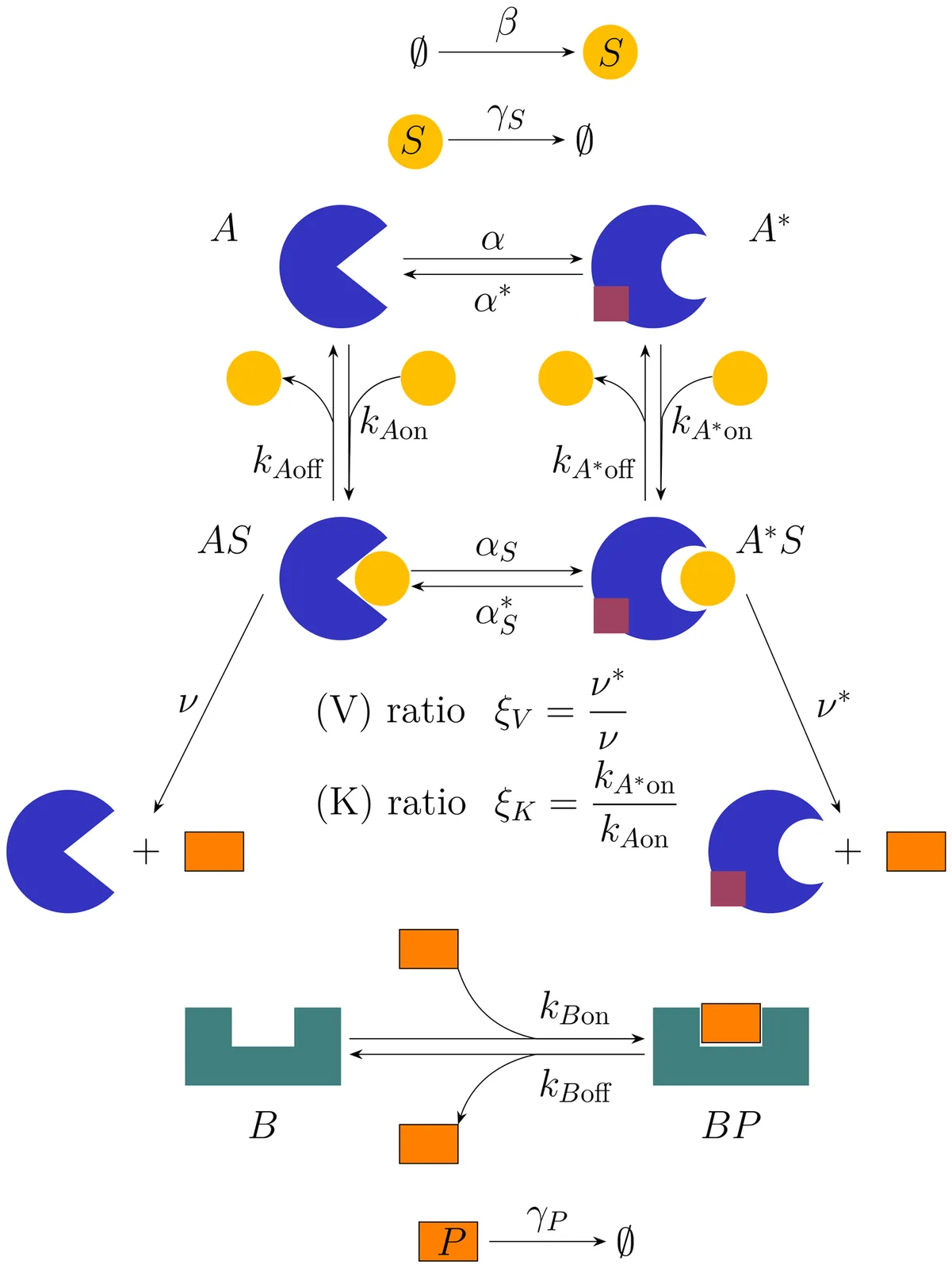
Allostery is a fundamental mechanism of protein regulation and is commonly interpreted as modulating enzymatic activity or product abundance. Here we show that this view is incomplete. Using a stochastic model of allosteric regulation combined with an information-theoretic analysis, we quantify the mutual information between an enzyme's regulatory state and the states of downstream signaling components. Beyond controlling steady-state production levels, allostery also regulates the timing and duration over which information is transmitted. By tuning the temporal operating regime of signaling pathways, allosteric regulation enables distinct dynamical outcomes from identical molecular components, providing a physical mechanism for temporal information flow, signaling specificity, and coordination without changes in metabolic pathways.
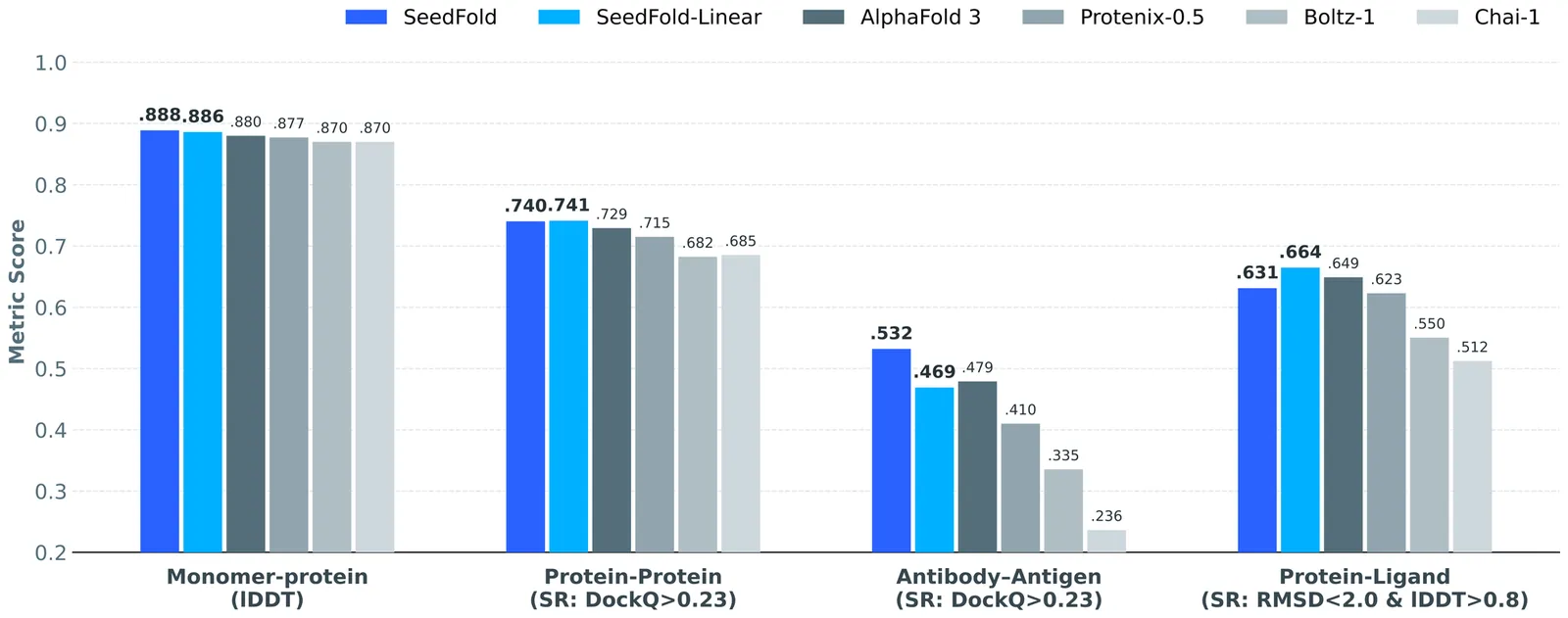
Highly accurate biomolecular structure prediction is a key component of developing biomolecular foundation models, and one of the most critical aspects of building foundation models is identifying the recipes for scaling the model. In this work, we present SeedFold, a folding model that successfully scales up the model capacity. Our contributions are threefold: first, we identify an effective width-scaling strategy for the Pairformer to increase representation capacity; second, we introduce a novel linear triangular attention that reduces computational complexity to enable efficient scaling; finally, we construct a large-scale distillation dataset to substantially enlarge the training set. Experiments on FoldBench show that SeedFold outperforms AlphaFold3 on most protein-related tasks.
Can machine learning models identify which chemist made a molecule from structure alone? If so, models trained on literature data may exploit chemist intent rather than learning causal structure-activity relationships. We test this by linking CHEMBL assays to publication authors and training a 1,815-class classifier to predict authors from molecular fingerprints, achieving 60% top-5 accuracy under scaffold-based splitting. We then train an activity model that receives only a protein identifier and an author-probability vector derived from structure, with no direct access to molecular descriptors. This author-only model achieves predictive power comparable to a simple baseline that has access to structure. This reveals a "Clever Hans" failure mode: models can predict bioactivity largely by inferring chemist goals and favorite targets without requiring a lab-independent understanding of chemistry. We analyze the sources of this leakage, propose author-disjoint splits, and recommend dataset practices to decouple chemist intent from biological outcomes.
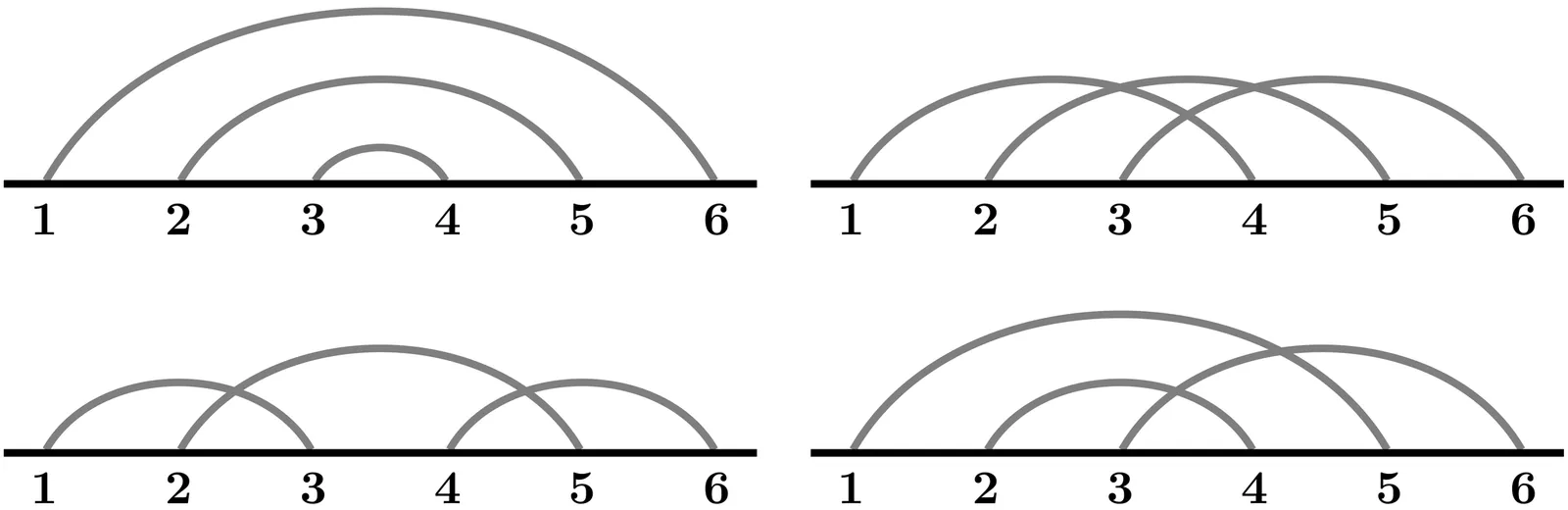
RNA molecules are known to form complex secondary structures including pseudoknots. A systematic framework for the enumeration, classification and prediction of secondary structures is critical to determine the biological significance of the molecular configurations of RNA. Chord diagrams are mathematical objects widely used to represent RNA secondary structures and to analyze structural motifs, however a mathematically rigorous enumeration of pseudoknots remains a challenge. We introduce a method that incorporates a distance-based metric $τ$ to analyze the intersection graph of a chord diagram associated with a pseudoknotted structure. In particular, our method formally defines a pseudoknot in terms of a weighted vertex cover of a certain intersection graph constructed from a partition of the chord diagram representing the nucleotide sequence of the RNA molecule. In this graph-theoretic context, we introduce a rigorous algorithm that enumerates pseudoknots, classifies secondary structures, and is sensitive to three-dimensional topological features. We implement our methods in MATLAB and test the algorithm on pseudoknotted structures from the bpRNA-1m database. Our findings confirm that genus is a robust quantifier of pseudoknot complexity.
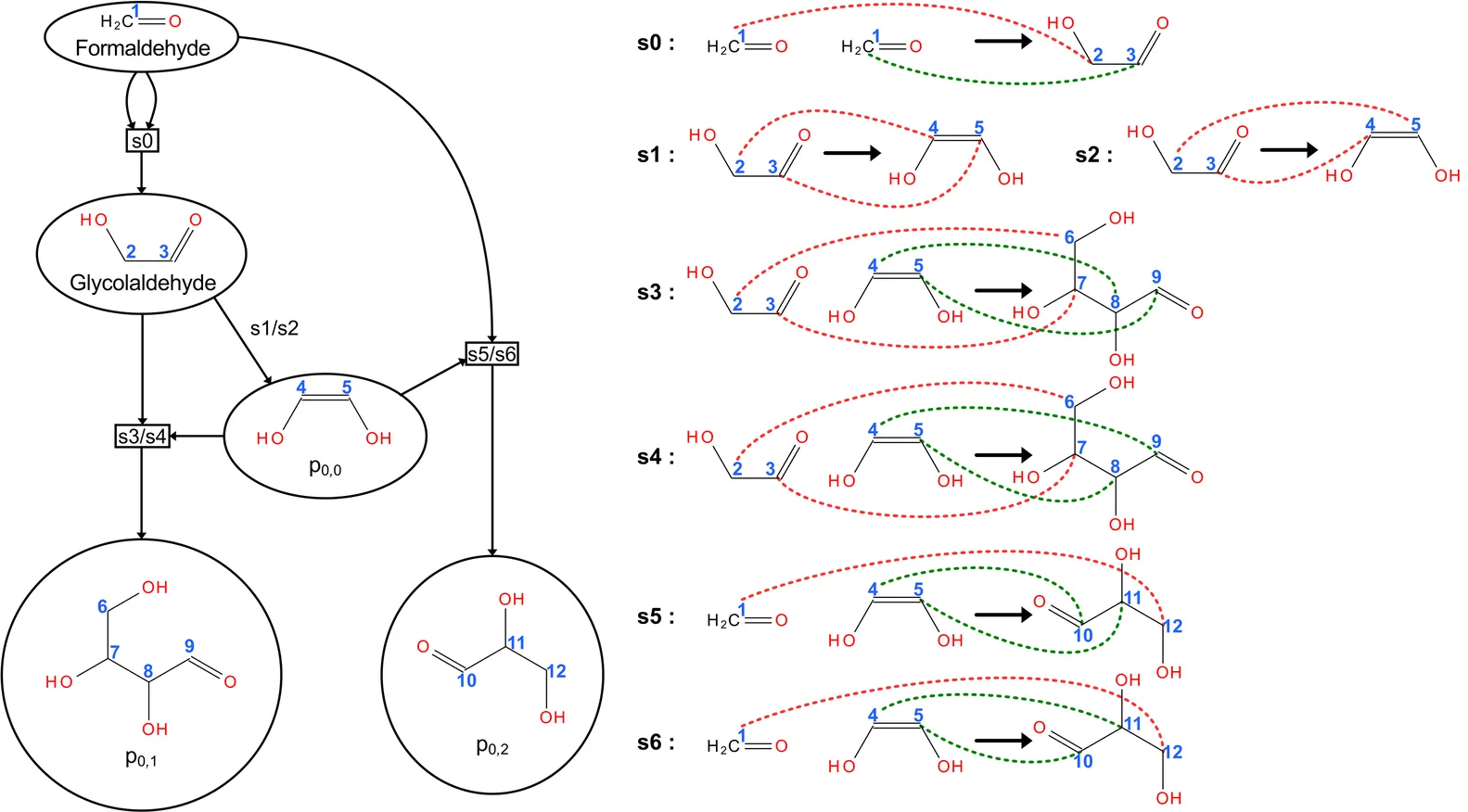
Atom tracing is essential for understanding the fate of labeled atoms in biochemical reaction networks, yet existing computational methods either simplify label correlations or suffer from combinatorial explosion. We introduce a saturation-based framework for enumerating labeling patterns that directly operates on atom-atom maps without requiring flux data or experimental measurements. The approach models reaction semantics using Kleisli morphisms in the powerset monad, allowing for compositional propagation of atom provenance through reaction networks. By iteratively saturating all possible educt combinations of reaction rules, the method exhaustively enumerates labeled molecular configurations, including multiplicities and reuse. Allowing arbitrary initial labeling patterns - including identical or distinct labels - the method expands only isotopomers reachable from these inputs, keeping the configuration space as small as necessary and avoids the full combinatorial growth characteristic of previous approaches. In principle, even every atom could carry a distinct identifier (e.g., tracing all carbon atoms individually), illustrating the generality of the framework beyond practical experimental limitations. The resulting template instance hypergraph captures the complete flow of atoms between compounds and supports projections tailored to experimental targets. Customizable labeling sets significantly reduce generated network sizes, providing efficient and exact atom traces focused on specific compounds or available isotopes. Applications to the tricarboxylic acid cycle, and glycolytic pathways demonstrate that the method fully automatically reproduces known labeling patterns and discovers steady-state labeling behavior. The framework offers a scalable, mechanistically transparent, and generalizable foundation for isotopomer modeling and experiment design.
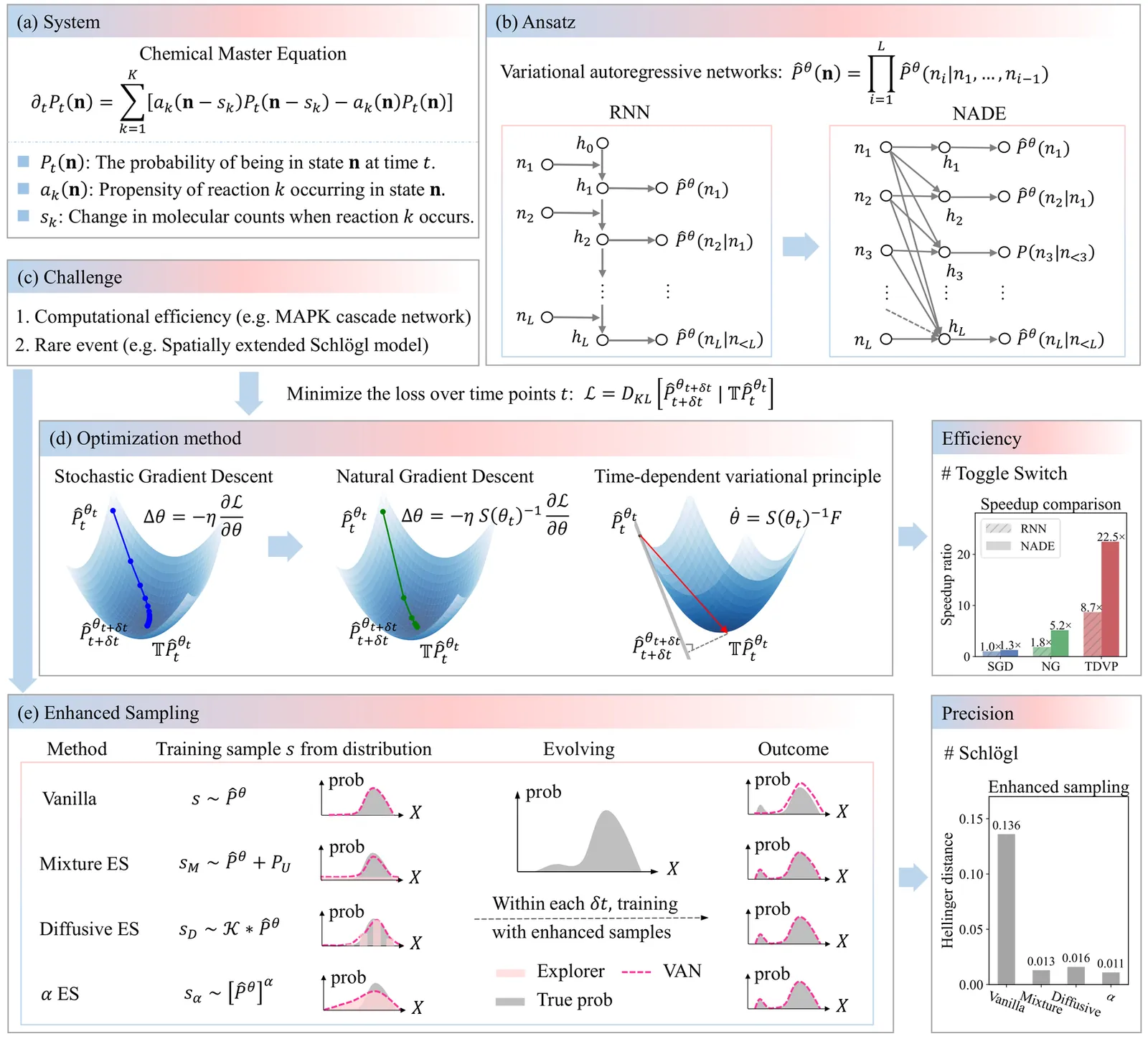
Chemical reaction networks are widely used to model stochastic dynamics in chemical kinetics, systems biology and epidemiology. Solving the chemical master equation that governs these systems poses a significant challenge due to the large state space exponentially growing with system sizes. The development of autoregressive neural networks offers a flexible framework for this problem; however, its efficiency is limited especially for high-dimensional systems and in scenarios with rare events. Here, we push the frontier of neural-network approach by exploiting faster optimizations such as natural gradient descent and time-dependent variational principle, achieving a 5- to 22-fold speedup, and by leveraging enhanced-sampling strategies to capture rare events. We demonstrate reduced computational cost and higher accuracy over the previous neural-network method in challenging reaction networks, including the mitogen-activated protein kinase (MAPK) cascade network, the hitherto largest biological network handled by the previous approaches of solving the chemical master equation. We further apply the approach to spatially extended reaction-diffusion systems, the Schlögl model with rare events, on two-dimensional lattices, beyond the recent tensor-network approach that handles one-dimensional lattices. The present approach thus enables efficient modeling of chemical reaction networks in general.
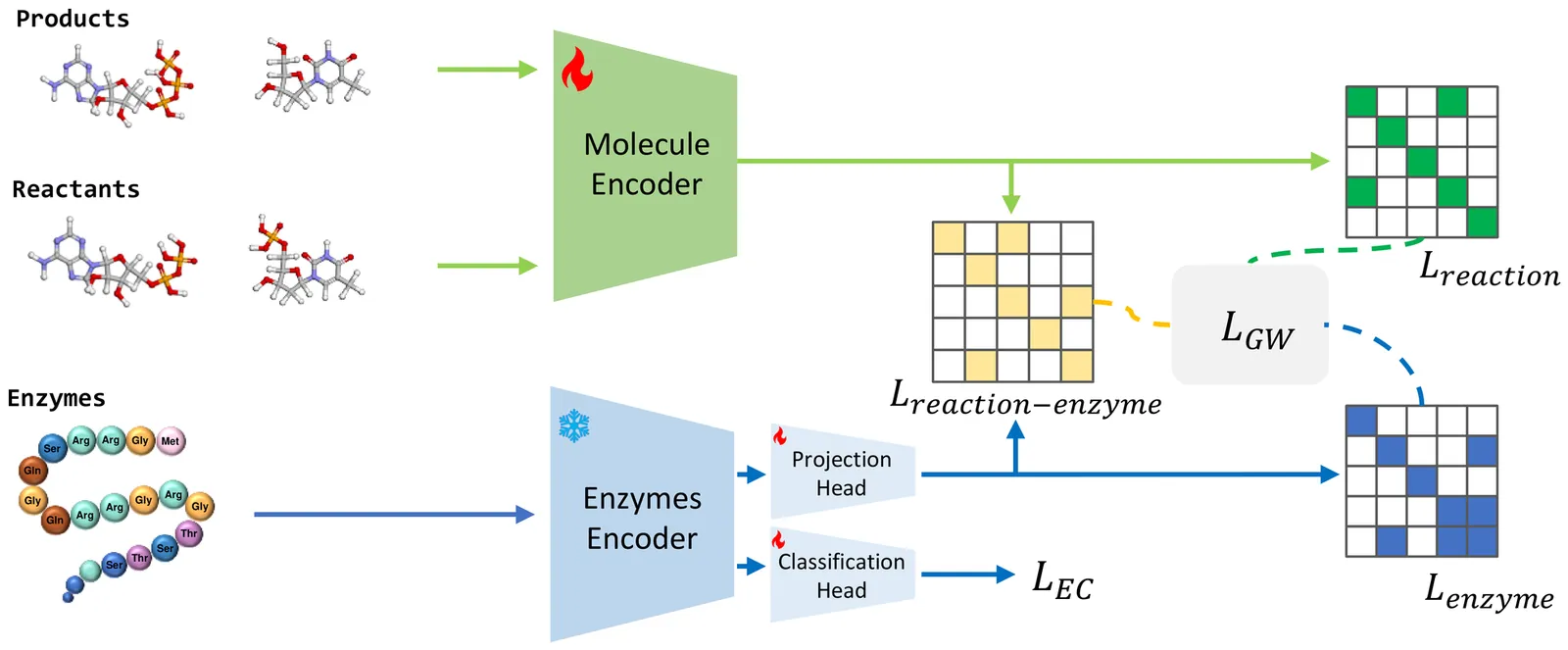
Enzymes are crucial catalysts that enable a wide range of biochemical reactions. Efficiently identifying specific enzymes from vast protein libraries is essential for advancing biocatalysis. Traditional computational methods for enzyme screening and retrieval are time-consuming and resource-intensive. Recently, deep learning approaches have shown promise. However, these methods focus solely on the interaction between enzymes and reactions, overlooking the inherent hierarchical relationships within each domain. To address these limitations, we introduce FGW-CLIP, a novel contrastive learning framework based on optimizing the fused Gromov-Wasserstein distance. FGW-CLIP incorporates multiple alignments, including inter-domain alignment between reactions and enzymes and intra-domain alignment within enzymes and reactions. By introducing a tailored regularization term, our method minimizes the Gromov-Wasserstein distance between enzyme and reaction spaces, which enhances information integration across these domains. Extensive evaluations demonstrate the superiority of FGW-CLIP in challenging enzyme-reaction tasks. On the widely-used EnzymeMap benchmark, FGW-CLIP achieves state-of-the-art performance in enzyme virtual screening, as measured by BEDROC and EF metrics. Moreover, FGW-CLIP consistently outperforms across all three splits of ReactZyme, the largest enzyme-reaction benchmark, demonstrating robust generalization to novel enzymes and reactions. These results position FGW-CLIP as a promising framework for enzyme discovery in complex biochemical settings, with strong adaptability across diverse screening scenarios.
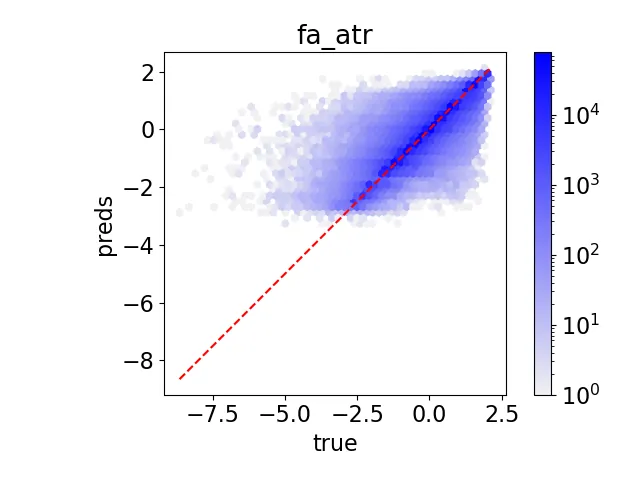
Understanding how protein mutations affect protein structure is essential for advancements in computational biology and bioinformatics. We introduce PRIMRose, a novel approach that predicts energy values for each residue given a mutated protein sequence. Unlike previous models that assess global energy shifts, our method analyzes the localized energetic impact of double amino acid insertions or deletions (InDels) at the individual residue level, enabling residue-specific insights into structural and functional disruption. We implement a Convolutional Neural Network architecture to predict the energy changes of each residue in a protein mutation. We train our model on datasets constructed from nine proteins, grouped into three categories: one set with exhaustive double InDel mutations, another with approximately 145k randomly sampled double InDel mutations, and a third with approximately 80k randomly sampled double InDel mutations. Our model achieves high predictive accuracy across a range of energy metrics as calculated by the Rosetta molecular modeling suite and reveals localized patterns that influence model performance, such as solvent accessibility and secondary structure context. This per-residue analysis offers new insights into the mutational tolerance of specific regions within proteins and provides higher interpretable and biologically meaningful predictions of InDels' effects.
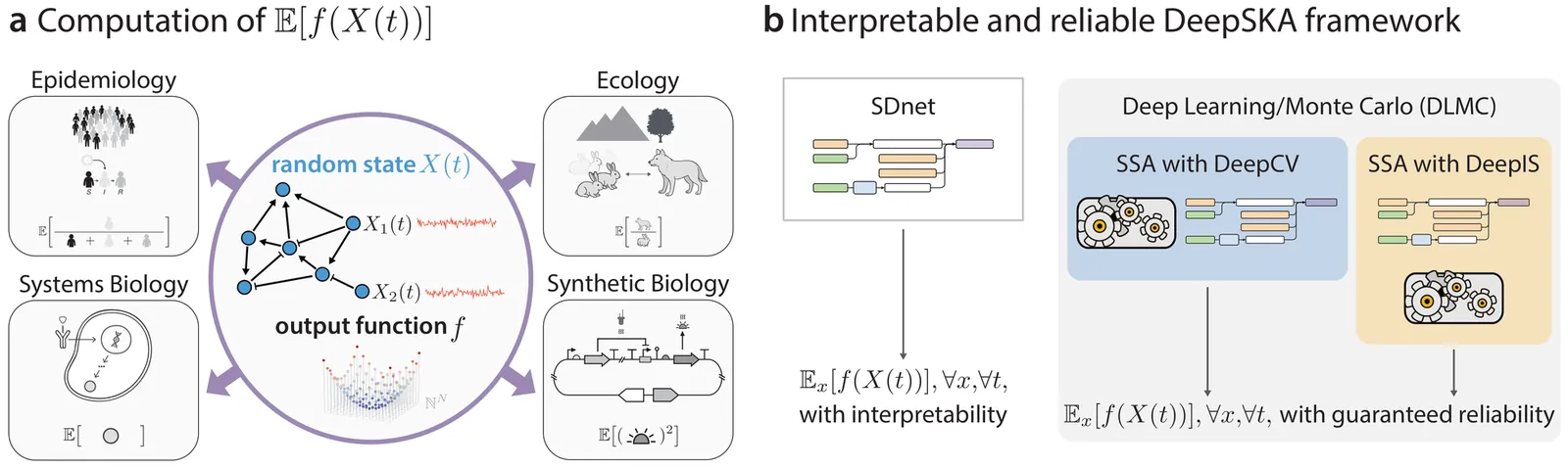
Stochastic Reaction Networks (SRNs) are a fundamental modeling framework for systems ranging from chemical kinetics and epidemiology to ecological and synthetic biological processes. A central computational challenge is the estimation of expected outputs across initial conditions and times, a task that is rarely solvable analytically and becomes computationally prohibitive with current methods such as Finite State Projection or the Stochastic Simulation Algorithm. Existing deep learning approaches offer empirical scalability, but provide neither interpretability nor reliability guarantees, limiting their use in scientific analysis and in applications where model outputs inform real-world decisions. Here we introduce DeepSKA, a neural framework that jointly achieves interpretability, guaranteed reliability, and substantial computational gains. DeepSKA yields mathematically transparent representations that generalise across states, times, and output functions, and it integrates this structure with a small number of stochastic simulations to produce unbiased, provably convergent, and dramatically lower-variance estimates than classical Monte Carlo. We demonstrate these capabilities across nine SRNs, including nonlinear and non-mass-action models with up to ten species, where DeepSKA delivers accurate predictions and orders-of-magnitude efficiency improvements. This interpretable and reliable neural framework offers a principled foundation for developing analogous methods for other Markovian systems, including stochastic differential equations.
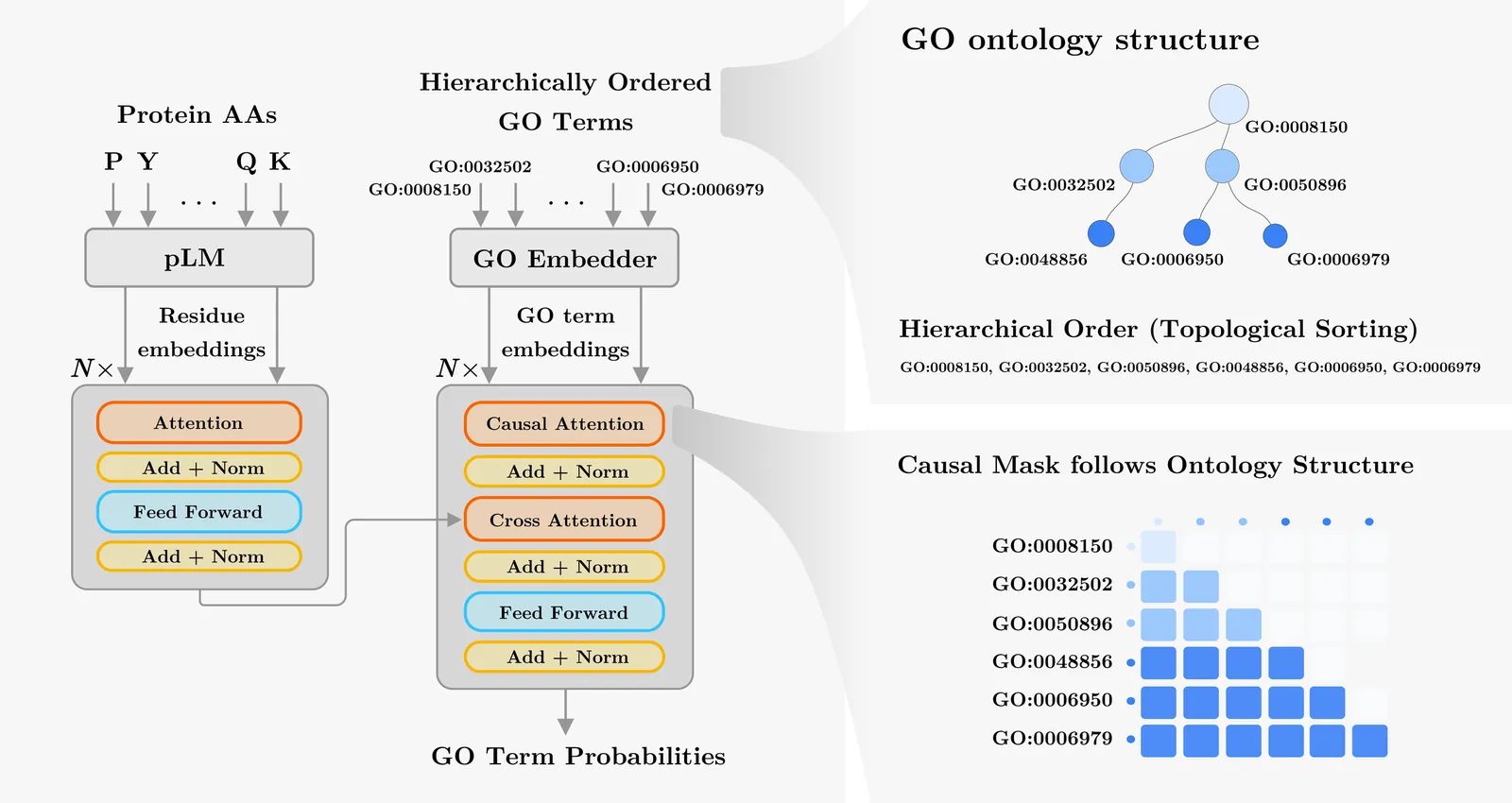
Accurate prediction of protein function is essential for elucidating molecular mechanisms and advancing biological and therapeutic discovery. Yet experimental annotation lags far behind the rapid growth of protein sequence data. Computational approaches address this gap by associating proteins with Gene Ontology (GO) terms, which encode functional knowledge through hierarchical relations and textual definitions. However, existing models often emphasize one modality over the other, limiting their ability to generalize, particularly to unseen or newly introduced GO terms that frequently arise as the ontology evolves, and making the previously trained models outdated. We present STAR-GO, a Transformer-based framework that jointly models the semantic and structural characteristics of GO terms to enhance zero-shot protein function prediction. STAR-GO integrates textual definitions with ontology graph structure to learn unified GO representations, which are processed in hierarchical order to propagate information from general to specific terms. These representations are then aligned with protein sequence embeddings to capture sequence-function relationships. STAR-GO achieves state-of-the-art performance and superior zero-shot generalization, demonstrating the utility of integrating semantics and structure for robust and adaptable protein function prediction. Code is available at https://github.com/boun-tabi-lifelu/stargo.
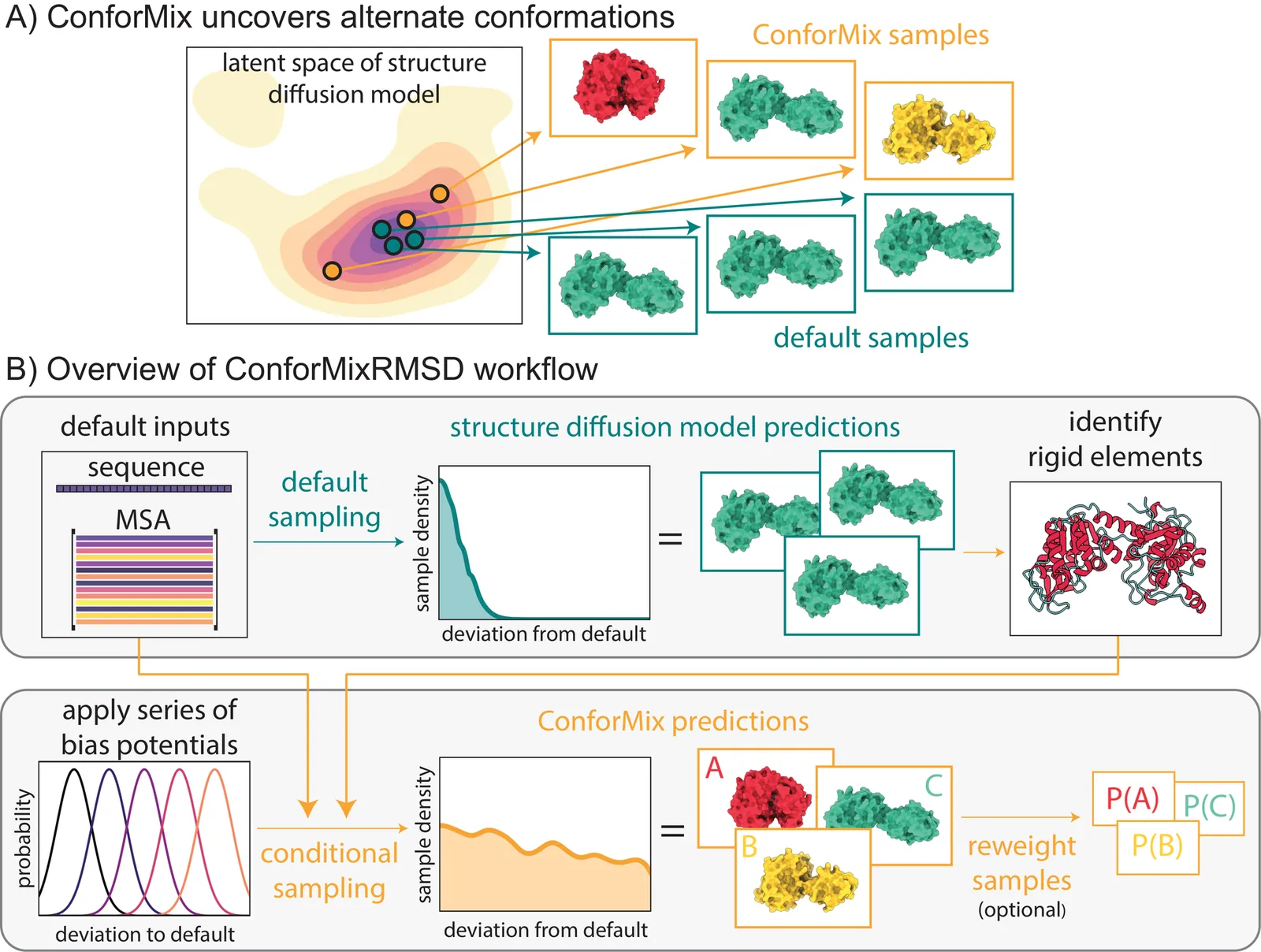
The function of biomolecules such as proteins depends on their ability to interconvert between a wide range of structures or "conformations." Researchers have endeavored for decades to develop computational methods to predict the distribution of conformations, which is far harder to determine experimentally than a static folded structure. We present ConforMix, an inference-time algorithm that enhances sampling of conformational distributions using a combination of classifier guidance, filtering, and free energy estimation. Our approach upgrades diffusion models -- whether trained for static structure prediction or conformational generation -- to enable more efficient discovery of conformational variability without requiring prior knowledge of major degrees of freedom. ConforMix is orthogonal to improvements in model pretraining and would benefit even a hypothetical model that perfectly reproduced the Boltzmann distribution. Remarkably, when applied to a diffusion model trained for static structure prediction, ConforMix captures structural changes including domain motion, cryptic pocket flexibility, and transporter cycling, while avoiding unphysical states. Case studies of biologically critical proteins demonstrate the scalability, accuracy, and utility of this method.
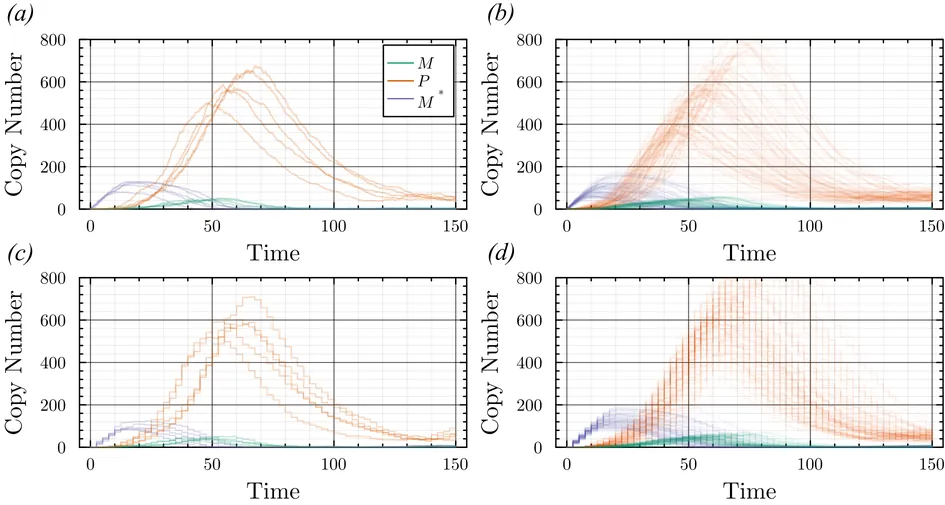
Stochastic models of biochemical reaction networks are widely used to capture intrinsic noise in cellular systems. The typical formulation of these models are based on Markov processes for which there is extensive research on efficient simulation and inference. However, there are biological processes, such as gene transcription and translation, that introduce history dependent dynamics requiring non-Markovian processes to accurately capture the stochastic dynamics of the system. This greater realism comes with additional computational challenges for simulation and parameter inference. We develop efficient stochastic simulation algorithms for well-mixed non-Markovian stochastic biochemical reaction networks with delays that depend on system state and time. Our methods generalize the next reaction method and $τ$-leaping method to support arbitrary inter-event time distributions while preserving computational scalability. We also introduce a coupling scheme to generate exact non-Markovian sample paths that are positively correlated to an approximate non-Markovian $τ$-leaping sample path. This enables substantial computational gains for Bayesian inference of model parameters though multifidelity simulation-based inference schemes. We demonstrate the effectiveness of our approach on a gene regulation model with delayed auto-inhibition, showing substantial gains in both simulation accuracy and inference efficiency of two orders of magnitude. These results extend the practical applicability of non-Markovian models in systems biology and beyond.
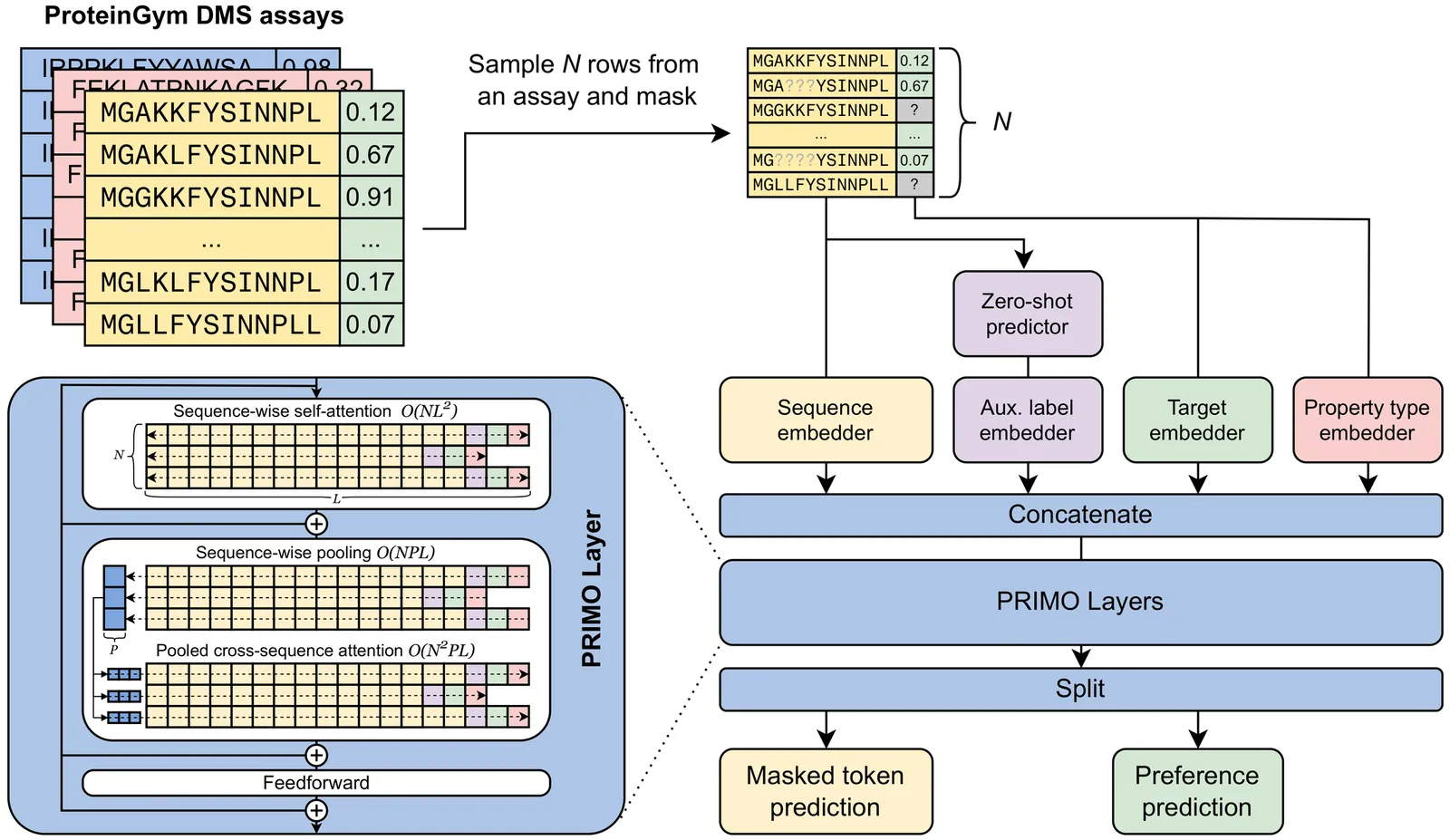
Accurately predicting protein fitness with minimal experimental data is a persistent challenge in protein engineering. We introduce PRIMO (PRotein In-context Mutation Oracle), a transformer-based framework that leverages in-context learning and test-time training to adapt rapidly to new proteins and assays without large task-specific datasets. By encoding sequence information, auxiliary zero-shot predictions, and sparse experimental labels from many assays as a unified token set in a pre-training masked-language modeling paradigm, PRIMO learns to prioritize promising variants through a preference-based loss function. Across diverse protein families and properties-including both substitution and indel mutations-PRIMO outperforms zero-shot and fully supervised baselines. This work underscores the power of combining large-scale pre-training with efficient test-time adaptation to tackle challenging protein design tasks where data collection is expensive and label availability is limited.
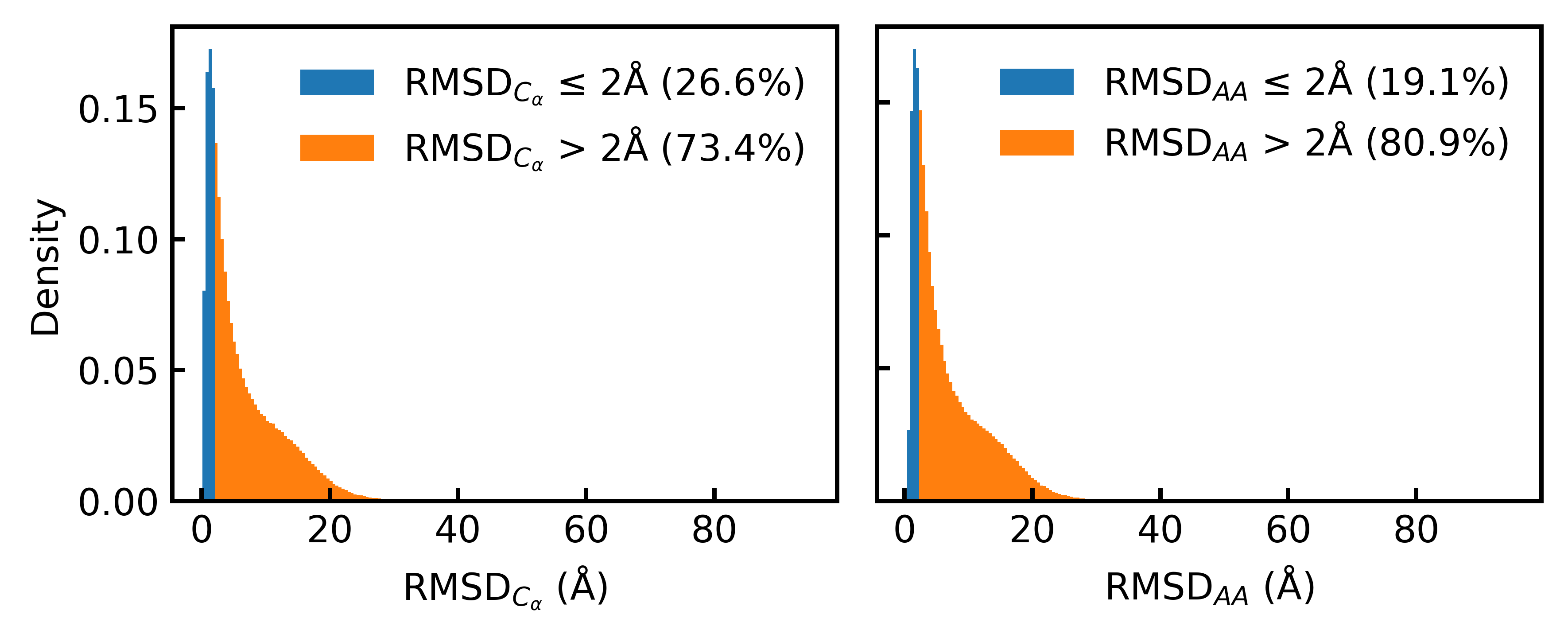
High-quality training datasets are crucial for the development of effective protein design models, but existing synthetic datasets often include unfavorable sequence-structure pairs, impairing generative model performance. We leverage ProteinMPNN, whose sequences are experimentally favorable as well as amenable to folding, together with structure prediction models to align high-quality synthetic structures with recoverable synthetic sequences. In that way, we create a new dataset designed specifically for training expressive, fully atomistic protein generators. By retraining La-Proteina, which models discrete residue type and side chain structure in a continuous latent space, on this dataset, we achieve new state-of-the-art results, with improvements of +54% in structural diversity and +27% in co-designability. To validate the broad utility of our approach, we further introduce Proteina Atomistica, a unified flow-based framework that jointly learns the distribution of protein backbone structure, discrete sequences, and atomistic side chains without latent variables. We again find that training on our new sequence-structure data dramatically boosts benchmark performance, improving \method's structural diversity by +73% and co-designability by +5%. Our work highlights the critical importance of aligned sequence-structure data for training high-performance de novo protein design models. All data will be publicly released.
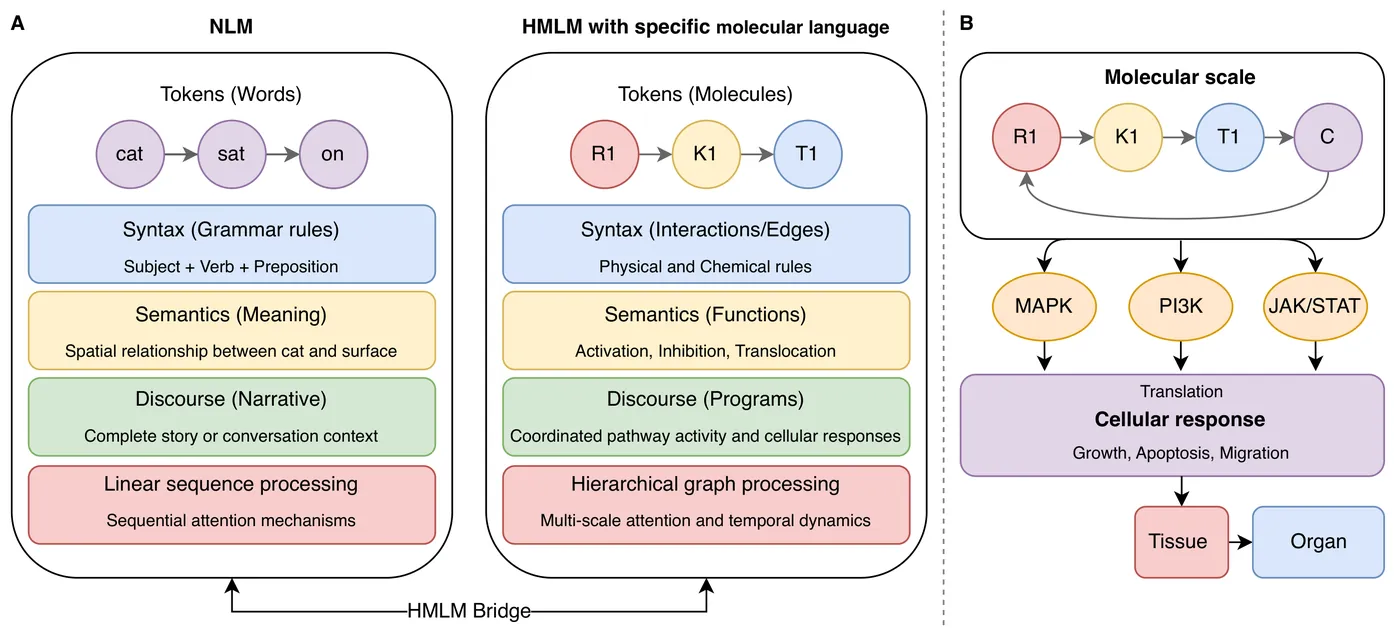
Artificial intelligence (AI) is reshaping computational and network biology by enabling new approaches to decode cellular communication networks. We introduce Hierarchical Molecular Language Models (HMLMs), a novel framework that models cellular signaling as a specialized molecular language, where signaling molecules function as tokens, protein interactions define syntax, and functional consequences constitute semantics. HMLMs employ a transformer-based architecture adapted to accommodate graph-structured signaling networks through information transducers, mathematical entities that capture how molecules receive, process, and transmit signals. The architecture integrates multi-modal data sources across molecular, pathway, and cellular scales through hierarchical attention mechanisms and scale-bridging operators that enable information flow across biological hierarchies. Applied to a complex network of cardiac fibroblast signaling, HMLMs outperformed traditional approaches in temporal dynamics prediction, particularly under sparse sampling conditions. Attention-based analysis revealed biologically meaningful crosstalk patterns, including previously uncharacterized interactions between signaling pathways. By bridging molecular mechanisms with cellular phenotypes through AI-driven molecular language representation, HMLMs establish a foundation for biology-oriented large language models (LLMs) that could be pre-trained on comprehensive pathway datasets and applied across diverse signaling systems and tissues, advancing precision medicine and therapeutic discovery.

Accurate prediction of enzyme kinetic parameters is crucial for drug discovery, metabolic engineering, and synthetic biology applications. Current computational approaches face limitations in capturing complex enzyme-substrate interactions and often focus on single parameters while neglecting the joint prediction of catalytic turnover numbers (Kcat) and Michaelis-Menten constants (Km). We present EnzyCLIP, a novel dual-encoder framework that leverages contrastive learning and cross-attention mechanisms to predict enzyme kinetic parameters from protein sequences and substrate molecular structures. Our approach integrates ESM-2 protein language model embeddings with ChemBERTa chemical representations through a CLIP-inspired architecture enhanced with bidirectional cross-attention for dynamic enzyme-substrate interaction modeling. EnzyCLIP combines InfoNCE contrastive loss with Huber regression loss to learn aligned multimodal representations while predicting log10-transformed kinetic parameters. The model is trained on the CatPred-DB database containing 23,151 Kcat and 41,174 Km experimentally validated measurements, and achieved competitive performance with R2 scores of 0.593 for Kcat and 0.607 for Km prediction. XGBoost ensemble methods applied to the learned embeddings further improved Km prediction (R2 = 0.61) while maintaining robust Kcat performance.
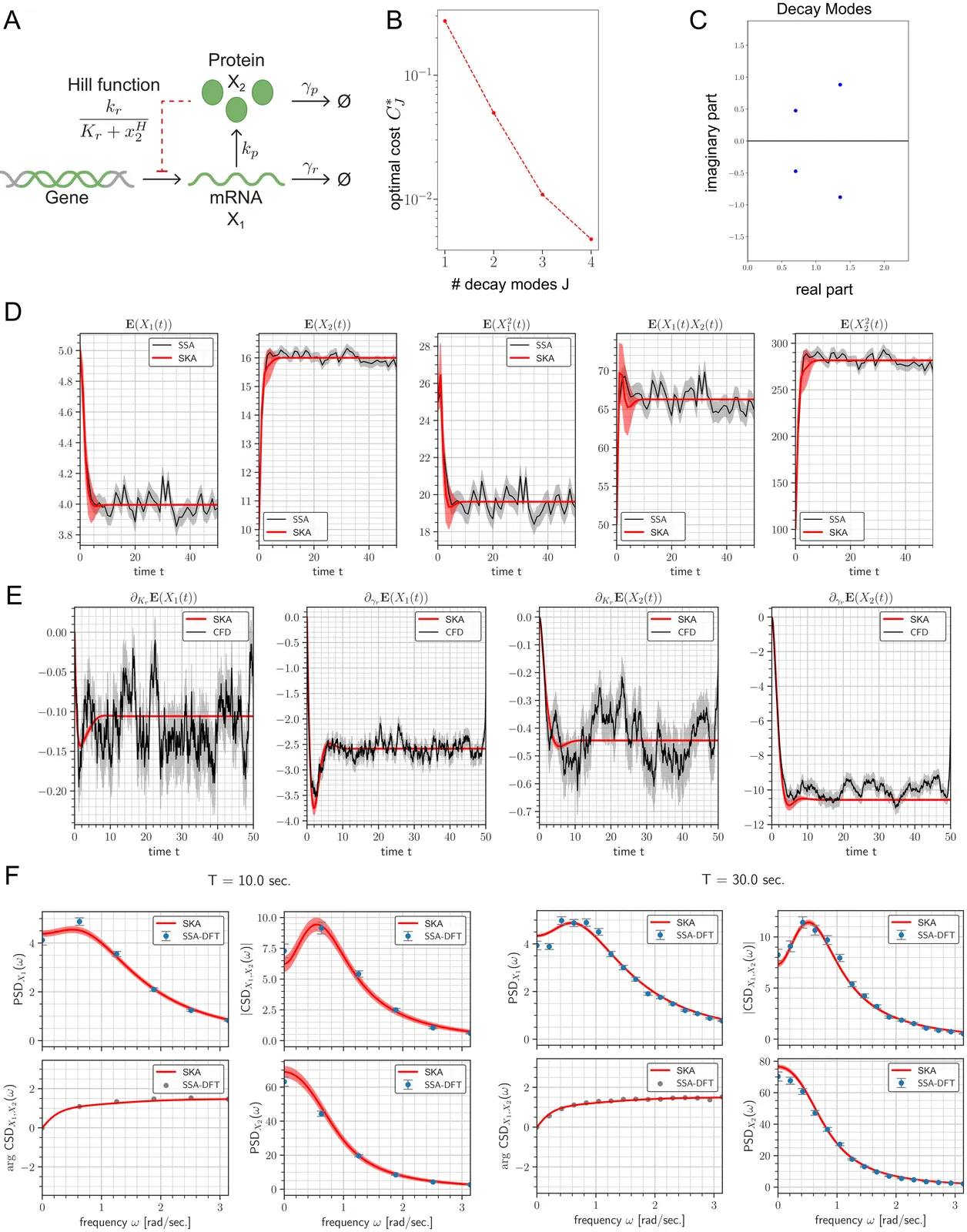
Stochastic reaction networks (SRNs) are a general class of continuous-time Markov jump processes used to model a wide range of systems, including biochemical dynamics in single cells, ecological and epidemiological populations, and queueing or communication networks. Yet analyzing their dynamics remains challenging because these processes are high-dimensional and their transient behavior can vary substantially across different initial molecular or population states. Here we introduce a spectral framework for the stochastic Koopman operator that provides a tractable, low-dimensional representation of SRN dynamics over continuous time, together with computable error estimates. By exploiting the compactness of the Koopman operator, we recover dominant spectral modes directly from simulated or experimental data, enabling efficient prediction of moments, event probabilities, and other summary statistics across all initial states. We further derive continuous-time parameter sensitivities and cross-spectral densities, offering new tools for probing noise structure and frequency-domain behavior. We demonstrate the approach on biologically relevant systems, including synthetic intracellular feedback controllers, stochastic oscillators, and inference of initial-state distributions from high-temporal-resolution flow cytometry. Together, these results establish spectral Koopman analysis as a powerful and general framework for studying stochastic dynamical systems across the biological, ecological, and computational sciences.